We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1706
From Proteopedia
(Difference between revisions)
| Line 43: | Line 43: | ||
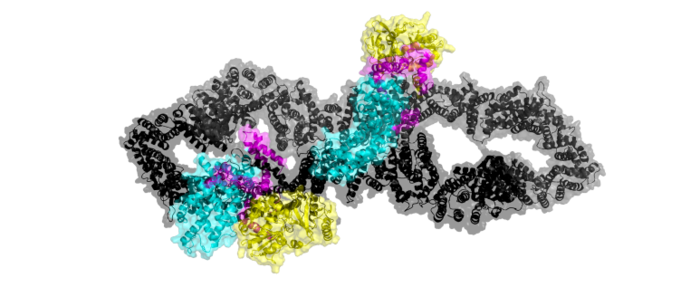
====Conformational Change Linkers==== | ====Conformational Change Linkers==== | ||
| - | These three helical linkers (L1, L2, and L3) undergo rotations to relocate the GRD and Sec14-PH sites | + | These three helical linkers (L1, L2, and L3) undergo rotations to relocate the GRD and Sec14-PH sites (</jmol> and <scene name='90/904311/Linkers_open/2'>linkers in open conformation</scene> |
<jmol> | <jmol> | ||
<jmolButton> | <jmolButton> | ||
| - | <script>moveto 1.0 { | + | <script>moveto 1.0 { 258 942 215 149.04} 254.03 0.0 0.0 {363.6198596926281 345.1996488441136 383.09000113650916} 143.7868028412415 {0 0 0} 0 0 0 3.0 0.0 0.0 </script> <text>🔎</text> |
</jmolButton> | </jmolButton> | ||
| - | </jmol> and <scene name='90/904311/ | + | </jmol> and <scene name='90/904311/Linker_closed/4'>linkers in closed conformation</scene> |
<jmol> | <jmol> | ||
<jmolButton> | <jmolButton> | ||
| - | <script>moveto 1.0 { | + | <script>moveto 1.0 { -93 -976 -198 100.01} 317.4 0.0 0.0 {364.4077734377054 341.9358671885079 392.3046403678749} 146.92471247967674 {0 0 0} 0 0 0 3.0 0.0 0.0 </script> <text>🔎</text> |
</jmolButton> | </jmolButton> | ||
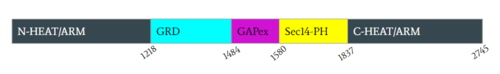
| - | </jmol>.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | + | </jmol>). Linker 1 (L1) consists of a loop connected by two helices from L1173-M1215 and is the main contributor in rotation of the GRD domain. The rotation of L1 to the open conformation causes N-HEAT/ARM [https://en.wikipedia.org/wiki/Alpha_helix α helix] 48 and GRD helix 49 to extend out, aligning to form a hinge point at G1190. The GRD relocation is assisted by Sec14-PH relocation, which is initiated by Linker 3(L3) from Q1835 to G1852. Movement of L3 is further supported by rearrangement of the proline rich section of the C-HEAT/ARM. L1 and L3 also move closer to each other in the open conformation initiating rearrangement of the GRD and Sec14-PH domain. Linker 2 (L2) consists of residues G1547-T1565 and begins at helix 63, the final helix of the GRD site, and connects into the short loop of [https://en.wikipedia.org/wiki/Alpha_helix α helix] 65 of the Sec14-PH domain and also assists in shifting the Sec14-PH away from the GRD site. The combination of these three linkers are largely responsible for the conformational shift of the closed and open conformation.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
==Ras Binding== | ==Ras Binding== | ||
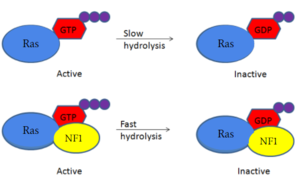
| - | Arg1276 is the critical residue within the GRD site for Ras activation. | + | Arg1276 is the critical residue within the GRD site for Ras activation. <scene name='90/904311/Arg_1276_open/13'>Arg 1276 in the open conformation</scene> |
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto 1.0 { -776 433 -458 146.98} 7209.11 0.0 0.0 {361.5375833333334 402.0207083333333 360.750125} 190.41144084626742 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> | ||
| + | </jmolButton> | ||
| + | </jmol>, the arginine finger binds to the backbone γ-carbon of Y32 in Ras to assist in eventual hydrolysis of GTP. <scene name='90/904311/Closed_zoom/10'>Arg 1276 in the closed conformation</scene> | ||
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto 1.0 { -816 -263 -515 73.84} 7673.3 0.0 0.0 {369.48317647058826 352.12982352941174 432.1719411764706} 186.8074765770222 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> | ||
| + | </jmolButton> | ||
| + | </jmol> interaction between Y32 and GTP is blocked by E31. Removal of the inhibition of E31 from Ras by R1276 from Neurofibromin allows for the normal function of neurofibromin in the rapid rate increase of GTP hydrolysis upon Ras binding. Mutations to the arginine finger slow GTPase activating reaction of neurofibromin.<ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
==SPRED 1== | ==SPRED 1== | ||
<scene name='90/904311/Spred1_w_grd/2'>SPRED 1</scene> is another peripheral protein that interacts with the GRD domain of Neurofibromin. SPRED 1 recruits Neurofibromin from the cytosol to the plasma membrane to interact with Ras. Binding of SPRED 1 can occur in either the open or closed conformation and causes a structural rearrangement of the GRD domain and GAPex subdomain that has yet to be structuralized.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | <scene name='90/904311/Spred1_w_grd/2'>SPRED 1</scene> is another peripheral protein that interacts with the GRD domain of Neurofibromin. SPRED 1 recruits Neurofibromin from the cytosol to the plasma membrane to interact with Ras. Binding of SPRED 1 can occur in either the open or closed conformation and causes a structural rearrangement of the GRD domain and GAPex subdomain that has yet to be structuralized.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
| Line 111: | Line 121: | ||
<scene name='90/904311/Arg_1276_open/14'>Arg 1276 w open zoomed out but with no Arg labeled</scene> | <scene name='90/904311/Arg_1276_open/14'>Arg 1276 w open zoomed out but with no Arg labeled</scene> | ||
| + | <scene name='90/904311/Arg_1276_open/13'>Arg 1276 in open conformation</scene> | ||
<jmol> | <jmol> | ||
<jmolButton> | <jmolButton> | ||
| - | <script>moveto 1.0 { -776 433 -458 146.98} 7209.11 0.0 0.0 {361.5375833333334 402.0207083333333 360.750125} 190.41144084626742 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text> | + | <script>moveto 1.0 { -776 433 -458 146.98} 7209.11 0.0 0.0 {361.5375833333334 402.0207083333333 360.750125} 190.41144084626742 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> |
</jmolButton> | </jmolButton> | ||
</jmol> | </jmol> | ||
| Line 131: | Line 142: | ||
<scene name='90/904311/Closed_zoom/11'>Arg 1276 closed zoomed out no label</scene> | <scene name='90/904311/Closed_zoom/11'>Arg 1276 closed zoomed out no label</scene> | ||
| - | + | <scene name='90/904311/Closed_zoom/10'>Arg 1276 in the closed conformation</scene> | |
<jmol> | <jmol> | ||
<jmolButton> | <jmolButton> | ||
| - | <script>moveto 1.0 { -816 -263 -515 73.84} 7673.3 0.0 0.0 {369.48317647058826 352.12982352941174 432.1719411764706} 186.8074765770222 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text> | + | <script>moveto 1.0 { -816 -263 -515 73.84} 7673.3 0.0 0.0 {369.48317647058826 352.12982352941174 432.1719411764706} 186.8074765770222 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> |
</jmolButton> | </jmolButton> | ||
</jmol> | </jmol> | ||
Revision as of 03:47, 19 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Neurofibromin 1
| |||||||||||