We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1706
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
===Domains=== | ===Domains=== | ||
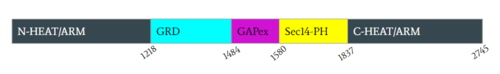
| - | [[Image:Domain.PNG|500px| | + | [[Image:Domain.PNG|500px|none|thumb|Figure 2: Domains for NF1 with their location on the <scene name='90/904311/Domainintro_open/11'>neurofibromin monomer</scene> and the pertinent colors. Numerical values show domain locations along the polypeptide chain.]] |
====N-C HEAT/ARM==== | ====N-C HEAT/ARM==== | ||
| Line 28: | Line 28: | ||
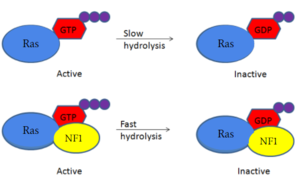
The <scene name='90/904311/Gapex_open/2'>GAPex</scene> of the GRD site lies between the Sec14-PH and GRD catalytic sites. This domain is non-catalytic and structurally consists of various loops and helices. Its may contribute to the binding of SPRED-1 to the GRD site. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | The <scene name='90/904311/Gapex_open/2'>GAPex</scene> of the GRD site lies between the Sec14-PH and GRD catalytic sites. This domain is non-catalytic and structurally consists of various loops and helices. Its may contribute to the binding of SPRED-1 to the GRD site. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
====Sec14-PH==== | ====Sec14-PH==== | ||
| - | The <scene name='90/904311/Sec14ph_open/2'>Sec14-PH </scene> is linked and extends out from the HEAT/ARM’s. Sec14-PH functions as a membrane associated domain and using a hydrophobic cavity binds to neurofibromin the plasma membrane. The | + | The <scene name='90/904311/Sec14ph_open/2'>Sec14-PH </scene> is linked and extends out from the HEAT/ARM’s. Sec14-PH functions as a membrane associated domain and using a hydrophobic cavity binds to neurofibromin the plasma membrane. The closed conformation also blocks Sec14-PH binding to the cell membrane due to blocking by GRD. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
===Conformations=== | ===Conformations=== | ||
====Closed conformation==== | ====Closed conformation==== | ||
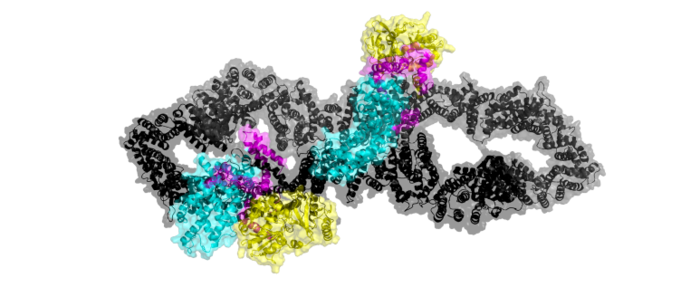
| - | Ras is unable to bind to the GRD binding site when both of the neurofibromin protomers are in | + | Ras is unable to bind to the GRD binding site when both of the neurofibromin protomers are in the <scene name='90/904311/Closed_confirmation_w_no_ras/3'>closed conformation</scene>. In the closed conformation, one protomer has its domains shifted by a 130° rotation of three separate conformational change linkers. That rotation places Arg1276 in an orientation where binding of Y32 of Ras is sterically hindered by E31. (<scene name='90/904311/Closed_arg/6'>Arg1276, Y32, and E31 interaction</scene> |
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto 1.0 { 347 936 -57 112.47} 2508.34 0.0 0.0 {366.9273333333333 357.0333333333333 434.887} 189.81815871562094 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> | ||
| + | </jmolButton> | ||
| + | </jmol>) Ras binding to the GRD site is inhibited by steric occlusion from the N-HEAT/ARM. The closed conformation can exist naturally without any form of stabilization but exists in a natural equilibrium with the open conformation.<ref name="Bourne"> DOI:10.1038/39470</ref> <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
=====Zinc Stabilized===== | =====Zinc Stabilized===== | ||
| - | The | + | The closed conformation is stabilized by a zinc ion that prevents the shift back to the open conformation. Zinc binding is chelated by three residues (<scene name='90/904311/Zinc_binding_site/11'>Cys1032, His1558, His1576</scene>). These three residues are contributed by the N-HEAT domain (Cys1032) and the GAPex-subdomain (His1558 and His1576). Zinc stabilization keeps Neurofibromin in the closed conformation, inhibiting Ras binding.<ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
====Open conformation==== | ====Open conformation==== | ||
| - | In the <scene name='90/904311/Open_conformation/14'> open conformation</scene>, one protomer is shifted to allow Ras binding, while the other protomer remains in the closed conformation. In the | + | In the <scene name='90/904311/Open_conformation/14'> open conformation</scene>, one protomer is shifted to allow Ras binding, while the other protomer remains in the closed conformation. In the open conformation one protomer is rotated 90°, facilitating binding between Y32 of RAS and Arg1276. (<scene name='90/904311/Arg_1276_mi/2'>Y32 and Arg1276 interaction</scene> |
| + | <jmol> | ||
| + | <jmolButton> | ||
| + | <script>moveto 1.0 { 294 -950 -102 45.37} 2089.03 0.0 0.0 {360.8673666666667 403.1638666666667 362.0271666666667} 189.97419116715565 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> | ||
| + | </jmolButton> | ||
| + | </jmol>) | ||
in the GRD site. To allow Ras binding, the GRD and Sec14-PH domains are reoriented away from one another and the GRD site is accessible for Ras binding. This conformational change to the open conformation is driven by rearrangement of three separate linkers (L1, L2, L3).<ref name="Bourne"> DOI:10.1038/39470</ref> <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | in the GRD site. To allow Ras binding, the GRD and Sec14-PH domains are reoriented away from one another and the GRD site is accessible for Ras binding. This conformational change to the open conformation is driven by rearrangement of three separate linkers (L1, L2, L3).<ref name="Bourne"> DOI:10.1038/39470</ref> <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
====Conformational Change Linkers==== | ====Conformational Change Linkers==== | ||
| - | These three helical linkers (L1, L2, and L3) undergo rotations to relocate the GRD and Sec14-PH sites ( | + | These three helical linkers (L1, L2, and L3) undergo rotations to relocate the GRD and Sec14-PH sites (<scene name='90/904311/Linkers_open/2'>linkers in open conformation</scene> |
<jmol> | <jmol> | ||
<jmolButton> | <jmolButton> | ||
| Line 80: | Line 90: | ||
Closed- | Closed- | ||
| - | <scene name='90/904311/Closed_confirmation_w_no_ras/3'> | + | <scene name='90/904311/Closed_confirmation_w_no_ras/3'>closed conformation</scene> |
Domain Scenes- | Domain Scenes- | ||
| Line 132: | Line 142: | ||
<jmol> | <jmol> | ||
<jmolButton> | <jmolButton> | ||
| - | <script>moveto 1.0 { 294 -950 -102 45.37} 2089.03 0.0 0.0 {360.8673666666667 403.1638666666667 362.0271666666667} 189.97419116715565 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text> | + | <script>moveto 1.0 { 294 -950 -102 45.37} 2089.03 0.0 0.0 {360.8673666666667 403.1638666666667 362.0271666666667} 189.97419116715565 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> |
</jmolButton> | </jmolButton> | ||
</jmol> | </jmol> | ||
| Line 154: | Line 164: | ||
<jmol> | <jmol> | ||
<jmolButton> | <jmolButton> | ||
| - | <script>moveto 1.0 { 347 936 -57 112.47} 2508.34 0.0 0.0 {366.9273333333333 357.0333333333333 434.887} 189.81815871562094 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text> | + | <script>moveto 1.0 { 347 936 -57 112.47} 2508.34 0.0 0.0 {366.9273333333333 357.0333333333333 434.887} 189.81815871562094 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> |
</jmolButton> | </jmolButton> | ||
</jmol> | </jmol> | ||
Revision as of 03:58, 19 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Neurofibromin 1
| |||||||||||