We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1706
From Proteopedia
(Difference between revisions)
| Line 84: | Line 84: | ||
<ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref><ref name="Sabatini"> DOI:10.1007/s11940-015-0355-4</ref> | <ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref><ref name="Sabatini"> DOI:10.1007/s11940-015-0355-4</ref> | ||
| - | ==FINAL SCENES COPY AND PASTE== | ||
| - | Open/Closed Conformation Scenes | ||
| - | |||
| - | |||
| - | Closed- | ||
| - | |||
| - | <scene name='90/904311/Closed_confirmation_w_no_ras/3'>closed conformation</scene> | ||
| - | |||
| - | Domain Scenes- | ||
| - | <scene name='90/904311/Domainintro_open/11'>Open conformation monomomer(paste at top of domain section)</scene> | ||
| - | |||
| - | <scene name='90/904311/N-heatarm_open/2'>N-HEATARM(place in heat section)</scene> | ||
| - | |||
| - | <scene name='90/904311/C-heatarm_open/2'>C-HEATARM(in heat section)</scene> | ||
| - | |||
| - | <scene name='90/904311/Grd_open/2'>GRD Open(in GRD section)</scene> | ||
| - | |||
| - | <scene name='90/904311/Gapex_open/2'>GAPex Open(in gap section)</scene> | ||
| - | |||
| - | <scene name='90/904311/Sec14ph_open/2'>Sec14PH Open(in sec section)</scene> | ||
| - | |||
| - | Linkers Open/Closed- | ||
| - | |||
| - | Closed Linker- | ||
| - | <scene name='90/904311/Linker_closed/4'>Linkers in Closed Conformation</scene> | ||
| - | <jmol> | ||
| - | <jmolButton> | ||
| - | <script>moveto 1.0 { -93 -976 -198 100.01} 317.4 0.0 0.0 {364.4077734377054 341.9358671885079 392.3046403678749} 146.92471247967674 {0 0 0} 0 0 0 3.0 0.0 0.0 </script> <text>🔎Zoom In(Closed Linkers)</text> | ||
| - | </jmolButton> | ||
| - | </jmol> | ||
| - | |||
| - | Open Linker- | ||
| - | <scene name='90/904311/Linkers_open/2'>Linkers in the Open Conformation</scene> | ||
| - | <jmol> | ||
| - | <jmolButton> | ||
| - | <script>moveto 1.0 { 258 942 215 149.04} 254.03 0.0 0.0 {363.6198596926281 345.1996488441136 383.09000113650916} 143.7868028412415 {0 0 0} 0 0 0 3.0 0.0 0.0 </script> <text>🔎Zoom in</text> | ||
| - | </jmolButton> | ||
| - | </jmol> | ||
| - | |||
| - | Zinc Binding Site Fixed- | ||
| - | <scene name='90/904311/Zinc_binding_site/11'>Zinc Binding Site</scene> | ||
| - | |||
| - | RAS Bound in Open Fixed- | ||
| - | |||
| - | <scene name='90/904311/Arg_1276_open/13'>Arg 1276 w open zoomed out</scene> | ||
| - | |||
| - | <scene name='90/904311/Arg_1276_open/14'>Arg 1276 w open zoomed out but with no Arg labeled</scene> | ||
| - | <scene name='90/904311/Arg_1276_open/13'>Arg 1276 in open conformation</scene> | ||
| - | <jmol> | ||
| - | <jmolButton> | ||
| - | <script>moveto 1.0 { -776 433 -458 146.98} 7209.11 0.0 0.0 {361.5375833333334 402.0207083333333 360.750125} 190.41144084626742 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> | ||
| - | </jmolButton> | ||
| - | </jmol> | ||
| - | |||
| - | Open Zoomed out Molecular Interactions: | ||
| - | <scene name='90/904311/Arg_1276_mi/2'>Molecular Interaction Open</scene> | ||
| - | <jmol> | ||
| - | <jmolButton> | ||
| - | <script>moveto 1.0 { 294 -950 -102 45.37} 2089.03 0.0 0.0 {360.8673666666667 403.1638666666667 362.0271666666667} 189.97419116715565 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> | ||
| - | </jmolButton> | ||
| - | </jmol> | ||
| - | |||
| - | RAS Bound in Closed Fixed- | ||
| - | |||
| - | <scene name='90/904311/Closed_zoom/10'>Arg 1276 closed zoomed out</scene> | ||
| - | |||
| - | <scene name='90/904311/Closed_zoom/11'>Arg 1276 closed zoomed out no label</scene> | ||
| - | |||
| - | <scene name='90/904311/Closed_zoom/10'>Arg 1276 in the closed conformation</scene> | ||
| - | <jmol> | ||
| - | <jmolButton> | ||
| - | <script>moveto 1.0 { -816 -263 -515 73.84} 7673.3 0.0 0.0 {369.48317647058826 352.12982352941174 432.1719411764706} 186.8074765770222 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> | ||
| - | </jmolButton> | ||
| - | </jmol> | ||
| - | |||
| - | Molecular Interactions closed R1276, E31, Y32: | ||
| - | |||
| - | <scene name='90/904311/Closed_arg/6'>Molecular Interactions R1276 Closed</scene> | ||
| - | <jmol> | ||
| - | <jmolButton> | ||
| - | <script>moveto 1.0 { 347 936 -57 112.47} 2508.34 0.0 0.0 {366.9273333333333 357.0333333333333 434.887} 189.81815871562094 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> | ||
| - | </jmolButton> | ||
| - | </jmol> | ||
| - | |||
| - | |||
| - | Spred1- | ||
| - | <scene name='90/904311/Spred1_w_grd/2'>Spred 1 closed w GRD</scene> | ||
| - | |||
| - | <scene name='90/904311/Closed_conformation/8'>Text To Be Displayed</scene> | ||
| - | |||
| - | |||
| - | ==References placeholders== | ||
| - | This section will be removed it is just for copy and paste purposes. We will add the PDB code names as references in the captions of each scene. | ||
| - | <ref name="Bergoug"> DOI:10.3390/cells9112365</ref> | ||
| - | <ref name="Bourne"> DOI:10.1038/39470</ref> | ||
| - | <ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref> | ||
| - | <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref> | ||
| - | <ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
| - | <ref name="Sabatini"> DOI:10.1007/s11940-015-0355-4</ref> | ||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 12:23, 19 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
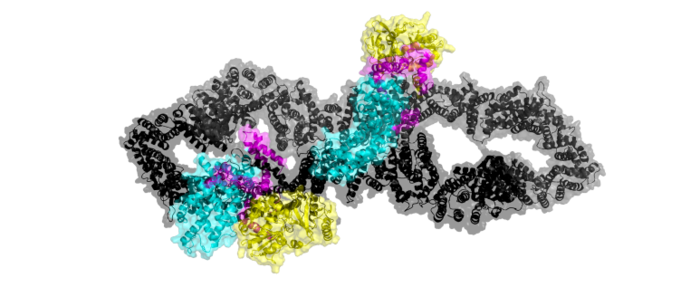
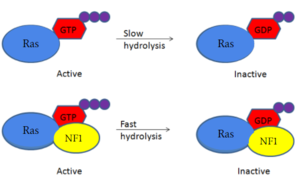
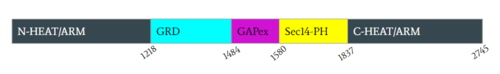
Neurofibromin 1
| |||||||||||