We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1722
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
MRGPRX2 also differs from typical Class A GPCRs based on substitutions in the PIF/connector motif, which acts as a microswitch. PIF plays a role in connecting the binding pocket to conformational rearrangements required for receptor activation<ref name="Schonegge">Schonegge, Anne-Marie, et al. "Evolutionary action and structural basis of the allosteric switch controlling β2AR functional selectivity." Nature, Nature Publishing Group, 18 December 2017, https://www.nature.com/articles/s41467-017-02257-x</ref>. The PIF motif is located towards the base of the TM domains. In a Class A GPCR, like <scene name='90/904327/B2arpif_pt_2/1'>β2AR, the PIF motif</scene> consists of Phe-211, Ile-121, and Phe-282 on TM domains 5, 3, and 6, respectively. However, for <scene name='90/904327/Mxpifnolabel_pt_3/1'>MRGPRX2, this PIF motif</scene> is replaced with Leu-194 on TM5, Leu-117 on TM3, and Phe-232 on TM6. These modifications result in tighter packing around the base of the receptor, indicating slightly different interactions with the intracellular [https://en.wikipedia.org/wiki/G_protein G-proteins]. | MRGPRX2 also differs from typical Class A GPCRs based on substitutions in the PIF/connector motif, which acts as a microswitch. PIF plays a role in connecting the binding pocket to conformational rearrangements required for receptor activation<ref name="Schonegge">Schonegge, Anne-Marie, et al. "Evolutionary action and structural basis of the allosteric switch controlling β2AR functional selectivity." Nature, Nature Publishing Group, 18 December 2017, https://www.nature.com/articles/s41467-017-02257-x</ref>. The PIF motif is located towards the base of the TM domains. In a Class A GPCR, like <scene name='90/904327/B2arpif_pt_2/1'>β2AR, the PIF motif</scene> consists of Phe-211, Ile-121, and Phe-282 on TM domains 5, 3, and 6, respectively. However, for <scene name='90/904327/Mxpifnolabel_pt_3/1'>MRGPRX2, this PIF motif</scene> is replaced with Leu-194 on TM5, Leu-117 on TM3, and Phe-232 on TM6. These modifications result in tighter packing around the base of the receptor, indicating slightly different interactions with the intracellular [https://en.wikipedia.org/wiki/G_protein G-proteins]. | ||
| - | [[Image:LabeledDRY.PNG|250px|right|thumb|'''Figure 2''': DRY Motif of MRGPRX2 <ref name="Yang">Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y</ref>]].(PDB entry:[https://www.rcsb.org/structure/7S8L 7S8L] | + | [[Image:LabeledDRY.PNG|250px|right|thumb|'''Figure 2''': DRY Motif of MRGPRX2 <ref name="Yang">Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y</ref>]].(PDB entry:[https://www.rcsb.org/structure/7S8L 7S8L]] |
| + | |||
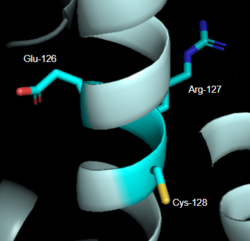
==== ''DRY Motif'' ==== | ==== ''DRY Motif'' ==== | ||
| Line 66: | Line 67: | ||
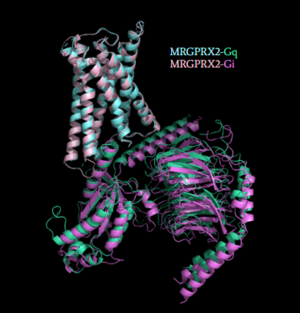
[[Image:Gq_and_Gi_snip.PNG|300px|right|thumb|'''Figure 5''': Comparison of the conformational change for MRGPRX2 (light blue) coupled with Gq (teal) and MRGPRX2 (pink) coupled with Gi (purple).<ref name="Cao"/> PDB entry [https://www.rcsb.org/structure/7S8N 7S8N] and [https://www.rcsb.org/structure/7S8O 7S8O]]] | [[Image:Gq_and_Gi_snip.PNG|300px|right|thumb|'''Figure 5''': Comparison of the conformational change for MRGPRX2 (light blue) coupled with Gq (teal) and MRGPRX2 (pink) coupled with Gi (purple).<ref name="Cao"/> PDB entry [https://www.rcsb.org/structure/7S8N 7S8N] and [https://www.rcsb.org/structure/7S8O 7S8O]]] | ||
| - | Once the ligand is bound, MRGPRX2 undergoes a conformational change that is transmitted through to the [https://en.wikipedia.org/wiki/G_protein#:~:text=G%20proteins%2C%20also%20known%20as,a%20cell%20to%20its%20interior. G-protein]. This conformational change is affected by the aforementioned deviances from Class A GPCRs. Rather than a large conformational change, a subtle one is induced. This will allow MRGPRX2 to interact with a G-protein. <scene name='90/904327/Gproteins/2'>G-proteins</scene> are composed of 3 subunits: α, β, and γ. When activated, the receptor acts as a [https://en.wikipedia.org/wiki/Guanine_nucleotide_exchange_factor Guanine nucleotide factor (GEF)]which will allow the Gα subunit to have its GDP be replaced by a GTP. This will cause the Gα subunit to dissociate from the dimer Gβγ <ref name="Ramesh">Ramesh, Soliman, et al. (2015) "G-Protein Coupled Receptors (GPCRs): A Comprehensive Computational Perspective." Combinational Chemistry and High Throughout Screening, 18(4), 346-364, https://pubmed.ncbi.nlm.nih.gov/25747435/</ref>. They are characterized by conserved motifs including DRY, PIF, Sodium Binding, and the CWxP <ref name="Zhou">PMID: 31855179</ref>. The Gα subunit is then able to act as a secondary messenger to begin the signal transduction in the cell. MRGPRX2 interacts with two different types of G-proteins; Gi and Gq. These G-proteins are activated by similar interactions with the receptor | + | Once the ligand is bound, MRGPRX2 undergoes a conformational change that is transmitted through to the [https://en.wikipedia.org/wiki/G_protein#:~:text=G%20proteins%2C%20also%20known%20as,a%20cell%20to%20its%20interior. G-protein]. This conformational change is affected by the aforementioned deviances from Class A GPCRs. Rather than a large conformational change, a subtle one is induced. This will allow MRGPRX2 to interact with a G-protein. <scene name='90/904327/Gproteins/2'>G-proteins</scene> are composed of 3 subunits: α, β, and γ. When activated, the receptor acts as a [https://en.wikipedia.org/wiki/Guanine_nucleotide_exchange_factor Guanine nucleotide factor (GEF)]which will allow the Gα subunit to have its GDP be replaced by a GTP. This will cause the Gα subunit to dissociate from the dimer Gβγ <ref name="Ramesh">Ramesh, Soliman, et al. (2015) "G-Protein Coupled Receptors (GPCRs): A Comprehensive Computational Perspective." Combinational Chemistry and High Throughout Screening, 18(4), 346-364, https://pubmed.ncbi.nlm.nih.gov/25747435/</ref>. They are characterized by conserved motifs including DRY, PIF, Sodium Binding, and the CWxP <ref name="Zhou">PMID: 31855179</ref>. The Gα subunit is then able to act as a secondary messenger to begin the signal transduction in the cell. MRGPRX2 interacts with two different types of G-proteins; Gi and Gq. These G-proteins are activated by similar interactions with the receptor due to their similar structures (Figure 5). |
=== 3.After G-Protein Activation === | === 3.After G-Protein Activation === | ||
| - | MRGPRX2 mediates degranulation of mast cells through interaction with Gq and Gi subunits. Gq activation signals through [https://en.wikipedia.org/wiki/Phospholipase_C Phospholipase C], which catalyzes the cleavage of Phosphatidylinositol bisphosphate into [https://en.wikipedia.org/wiki/Diglyceride diacylglycerol (DAG)] and [https://en.wikipedia.org/wiki/Inositol_trisphosphate inositol trisphosphate (IP3)]. DAG is then able to increase the activity of [https://en.wikipedia.org/wiki/Protein_kinase_C protein kinase C]. IP3 receptor is ligand gated calcium channel on the endoplasmic reticulum (ER), that causes the | + | MRGPRX2 mediates degranulation of mast cells through interaction with Gq and Gi subunits. Gq activation signals through [https://en.wikipedia.org/wiki/Phospholipase_C Phospholipase C], which catalyzes the cleavage of Phosphatidylinositol bisphosphate into [https://en.wikipedia.org/wiki/Diglyceride diacylglycerol (DAG)] and [https://en.wikipedia.org/wiki/Inositol_trisphosphate inositol trisphosphate (IP3)]. DAG is then able to increase the activity of [https://en.wikipedia.org/wiki/Protein_kinase_C protein kinase C]. IP3 receptor is ligand gated calcium channel on the endoplasmic reticulum (ER), that causes the release of calcium in cytoplasm. Subsequently, this results in muscle contraction and enzyme activation. |
| - | MRGPRX2 also interacts with Gi or G-protein inhibitory, which is structurally homologous to Gs (G-protein stimulatory) | + | MRGPRX2 also interacts with Gi or G-protein inhibitory, which is structurally homologous to Gs (G-protein stimulatory) and will inhibit adenylyl cyclase and therefore lowers (cAMP). Gi is stimulated when [https://en.wikipedia.org/wiki/Somatostatin somatostatin] binds to receptor in pancreas. |
== Clinical Relevance == | == Clinical Relevance == | ||
| Line 78: | Line 79: | ||
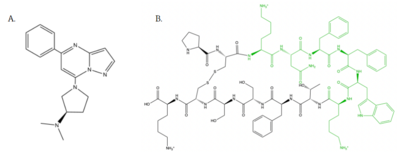
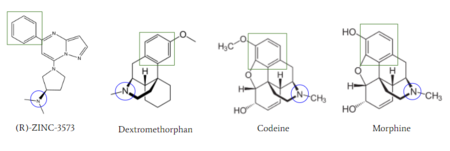
[[Image:Zinc_and_drugs_snip.PNG|450px|right|thumb|'''Figure 6''': Structures of (R)-Zinc-3573, Dextromethorphan, Morphine, and Codeine. The blue circles indicate conserved basic N-dimethyl group and the green squares show the conserved benzene rings.<ref name="Cao"/>]] | [[Image:Zinc_and_drugs_snip.PNG|450px|right|thumb|'''Figure 6''': Structures of (R)-Zinc-3573, Dextromethorphan, Morphine, and Codeine. The blue circles indicate conserved basic N-dimethyl group and the green squares show the conserved benzene rings.<ref name="Cao"/>]] | ||
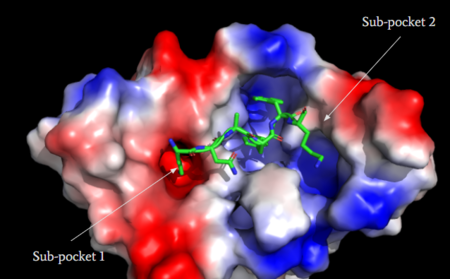
| - | MRGPRX2 activation is associated with chronic itching or anaphylaxis reactions, a common side effect of many prescribed medications. <ref name="Cao"/> Among these drugs are | + | MRGPRX2 activation is associated with chronic itching or anaphylaxis reactions, a common side effect of many prescribed medications. <ref name="Cao"/> Among these drugs are opioids [https://en.wikipedia.org/wiki/Morphine morphine] and [https://en.wikipedia.org/wiki/Codeine codeine] and [https://en.wikipedia.org/wiki/Dextromethorphan dextromethorphan]. By analyzing the structures of these drugs, it can be seen that they contain chemical features similar to <scene name='90/904328/Zizwithaa/5'>agonist R-Zinc-3573</scene> (see Figure 6). They contain a conserved benzene ring that stabilizes them in binding pocket one (see Figure 6). Similarly, they contain an N-dimethyl group that would allow them to form key ionic bonds with residues Asp-184 and Glu-164. Lastly, these drugs have a similar shape and size to that of the agonist R-Zinc-3573 <ref name="Babina"> Babina, M., et al. "MRGPRX2 Is the Codeine Receptor of Human Skin Mast Cells: Desensitization through β-Arrestin and Lack of Correlation with the FcεRI Pathway." Journal of Investigative Dermatology, 141(6), 1286-1296. https://doi.org/10.1016/j.jid.2020.09.017</ref>. These structural and chemical similarities indicate the possibility of a similar binding mechanism to previously discussed agonist, (R)-Zinc-3573. |
| - | + | ||
| - | + | ||
| - | </ | + | This receptor is shallow and is able to bind to a variety of drugs, creating a need for a solution to mediate the effects of MRGPRX2 activation. Currently, there is research for the development of small antagonists that will induce an anti-inflammatory effect by preventing [https://en.wikipedia.org/wiki/Immunoglobulin_E IgE-dependent] mast cell degranulation <ref name="McNeil">McNeil, B. D., et al. "MRGPRX2 and Adverse Drug Reactions." Frontier Immunology, 06 August 2021, https://www.frontiersin.org/articles/10.3389/fimmu.2021.676354/full</ref>. There are two antagonists that have been adapted from being antagonists to another Class A GPCR, [https://en.wikipedia.org/wiki/NK1_receptor_antagonist neurokinin-1 receptor (NK-1R)] <ref name="Ogasawara">Ogasawara, H., et al. "Novel MRGPRX2 antagonists inhibit IgE-independent activation of human umbilical cord blood-derived mast cells." Journal of Leukocyte Biology, 12 July 2019, https://jlb.onlinelibrary.wiley.com/doi/10.1002/JLB.2AB1018-405R</ref>. Through the development of this research, these adverse side effects can be relieved and improve patient satisfaction. |
| - | = | + | |
| - | < | + | |
Revision as of 13:47, 21 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Human Itch Mas-Related G-Protein Coupled Receptor
| |||||||||||