This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Neurofibromin
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
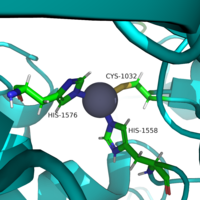
The Gap-related domain, or <scene name='90/904326/Grd_highlighted/1'>GRD</scene>, is the catalytic domain of neurofibromin. It ranges from residues 1196 to 1547. <ref name="Naschberger"/> Its main catalytic mechanism is the hydrolysis of GTP-bound Ras into GDP-bound Ras, which converts Ras from its active form into its inactive form. The GRD provides an arginine residue, known as the arginine finger, to Ras. The location of the Gap-related domain is shifted between the <scene name='90/904326/Grdopen/1'>open conformation</scene> and closed conformations of neurofibromin. | The Gap-related domain, or <scene name='90/904326/Grd_highlighted/1'>GRD</scene>, is the catalytic domain of neurofibromin. It ranges from residues 1196 to 1547. <ref name="Naschberger"/> Its main catalytic mechanism is the hydrolysis of GTP-bound Ras into GDP-bound Ras, which converts Ras from its active form into its inactive form. The GRD provides an arginine residue, known as the arginine finger, to Ras. The location of the Gap-related domain is shifted between the <scene name='90/904326/Grdopen/1'>open conformation</scene> and closed conformations of neurofibromin. | ||
==== Sec-PH ==== | ==== Sec-PH ==== | ||
| - | The <scene name='90/ | + | The <scene name='90/904325/Secph_highlighted/2'>Sec14-PH</scene> domain is the lipid-binding domain of neurofibromin, found in residues 1565 to 1835. <ref name="Naschberger"/> In the closed conformation of neurofibromin, the hydrophobic core is blocked by the Gap-related domain. The <scene name='90/904326/Secopen/1'>open conformation</scene> allows the hydrophobic core in the Sec cavity to be accessible and exposed. In neurofibromin, this cavity binds [https://en.wikipedia.org/wiki/Glycerophospholipid glycerophospholipids], which can induce conformational changes. <ref>DOI 10.1016/j.febslet.2012.06.006</ref> It is unclear if the Sec14-PH domain has a role in the RasGap activity of neurofibromin. |
===Important Structural Features=== | ===Important Structural Features=== | ||
====Conformations==== | ====Conformations==== | ||
Revision as of 17:02, 21 April 2022
| |||||||||||
References
- ↑ 1.0 1.1 Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Naschberger A, Baradaran R, Rupp B, Carroni M. The structure of neurofibromin isoform 2 reveals different functional states. Nature. 2021 Nov;599(7884):315-319. doi: 10.1038/s41586-021-04024-x. Epub 2021, Oct 27. PMID:34707296 doi:http://dx.doi.org/10.1038/s41586-021-04024-x
- ↑ Trovo-Marqui AB, Tajara EH. Neurofibromin: a general outlook. Clin Genet. 2006 Jul;70(1):1-13. doi: 10.1111/j.1399-0004.2006.00639.x. PMID:16813595 doi:http://dx.doi.org/10.1111/j.1399-0004.2006.00639.x
- ↑ Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12138-42. Epub 2002 Sep 4. PMID:12213964 doi:http://dx.doi.org/10.1073/pnas.192453199
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Yoshimura SH, Hirano T. HEAT repeats - versatile arrays of amphiphilic helices working in crowded environments? J Cell Sci. 2016 Nov 1;129(21):3963-3970. doi: 10.1242/jcs.185710. Epub 2016 Oct , 6. PMID:27802131 doi:http://dx.doi.org/10.1242/jcs.185710
- ↑ 7.0 7.1 7.2 Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2
- ↑ Scheffzek K, Welti S. Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett. 2012 Aug 14;586(17):2662-73. doi: 10.1016/j.febslet.2012.06.006. Epub , 2012 Jun 19. PMID:22728242 doi:http://dx.doi.org/10.1016/j.febslet.2012.06.006
- ↑ Dunzendorfer-Matt T, Mercado EL, Maly K, McCormick F, Scheffzek K. The neurofibromin recruitment factor Spred1 binds to the GAP related domain without affecting Ras inactivation. Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):7497-502. doi:, 10.1073/pnas.1607298113. Epub 2016 Jun 16. PMID:27313208 doi:http://dx.doi.org/10.1073/pnas.1607298113
- ↑ Frech M, Darden TA, Pedersen LG, Foley CK, Charifson PS, Anderson MW, Wittinghofer A. Role of glutamine-61 in the hydrolysis of GTP by p21H-ras: an experimental and theoretical study. Biochemistry. 1994 Mar 22;33(11):3237-44. doi: 10.1021/bi00177a014. PMID:8136358 doi:http://dx.doi.org/10.1021/bi00177a014
- ↑ Bunda S, Burrell K, Heir P, Zeng L, Alamsahebpour A, Kano Y, Raught B, Zhang ZY, Zadeh G, Ohh M. Inhibition of SHP2-mediated dephosphorylation of Ras suppresses oncogenesis. Nat Commun. 2015 Nov 30;6:8859. doi: 10.1038/ncomms9859. PMID:26617336 doi:http://dx.doi.org/10.1038/ncomms9859
- ↑ Abramowicz A, Gos M. Neurofibromin in neurofibromatosis type 1 - mutations in NF1gene as a cause of disease. Dev Period Med. 2014 Jul-Sep;18(3):297-306. PMID:25182393
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Ly KI, Blakeley JO. The Diagnosis and Management of Neurofibromatosis Type 1. Med Clin North Am. 2019 Nov;103(6):1035-1054. doi: 10.1016/j.mcna.2019.07.004. PMID:31582003 doi:http://dx.doi.org/10.1016/j.mcna.2019.07.004
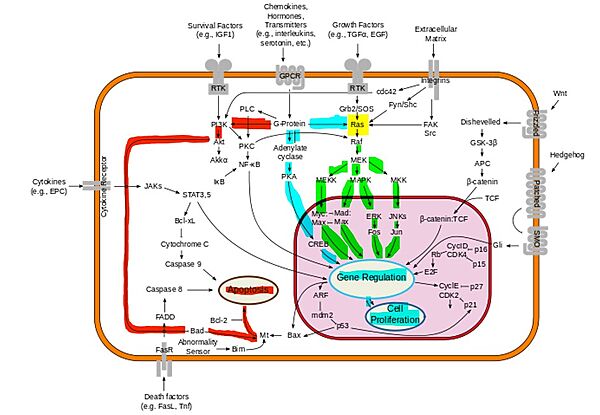
- ↑ McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, Stivala F, Libra M, Basecke J, Evangelisti C, Martelli AM, Franklin RA. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007 Aug;1773(8):1263-84. doi:, 10.1016/j.bbamcr.2006.10.001. Epub 2006 Oct 7. PMID:17126425 doi:http://dx.doi.org/10.1016/j.bbamcr.2006.10.001
Proteopedia Page Contributors and Editors (what is this?)
Jordyn K. Lenard, Ryan D. Adkins, Michal Harel, OCA, Jaime Prilusky