This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Amer Ali/Sandbox 2
From Proteopedia
| Line 8: | Line 8: | ||
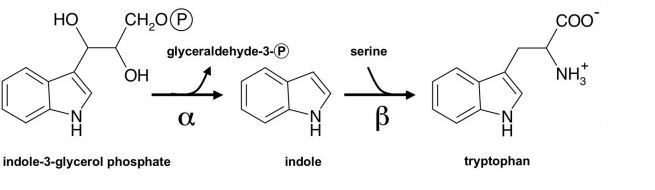
TrpS is a pyridoxal 5’-phosphate (PLP)-dependent enzyme. TrpS has an alpha and beta chain that form a linear alpha-beta-beta-alpha heterotetrameric complex <ref name= "Michalska"> PMID: 31316809 </ref>. This is termed as the alpha2beta2 complex, and each subunit is also referred to as TrpA and TrpB for the alpha and beta subunits respectively. The alpha active site contains catalytic residues Glu and Asp, and a hydrophobic intramolecular tunnel allows for the transport of indole from the alpha subunit active site to the beta subunit active site (Miles, 2001). The alpha and beta chain are encoded by trpA and trpB genes that are involved in TrpS regulatory operon. In bacteria and plants, the alpha and beta chains are separate, but in fungi the two chains are fused into one protein (Tryptophan synthase, alpha chain, active site). The alpha2beta2 tetramer complex contains pyridoxal-phosphate, glycerol-3-phosphate, Na+ ions. Another key site in tryptophan synthase is the monovalent cation (MVC) site, which is made up of cations like Na+ and K+, along with Cs+ <ref name= "Dierkers"> DOI: 10.1021/bi9008374</ref>. For regulation, TrpA and TrpB cycle between low-activity open conformation and a high-activity closed state. This is done by the binding of an IGP substrate to TrpA which promotes the high activity closed state which activates the TrpB high activity closed state. The states are reset back to low activity open conformation by the production of L-Trp external aldimine <ref name= "Michalska"> PMID: 31316809 </ref>. | TrpS is a pyridoxal 5’-phosphate (PLP)-dependent enzyme. TrpS has an alpha and beta chain that form a linear alpha-beta-beta-alpha heterotetrameric complex <ref name= "Michalska"> PMID: 31316809 </ref>. This is termed as the alpha2beta2 complex, and each subunit is also referred to as TrpA and TrpB for the alpha and beta subunits respectively. The alpha active site contains catalytic residues Glu and Asp, and a hydrophobic intramolecular tunnel allows for the transport of indole from the alpha subunit active site to the beta subunit active site (Miles, 2001). The alpha and beta chain are encoded by trpA and trpB genes that are involved in TrpS regulatory operon. In bacteria and plants, the alpha and beta chains are separate, but in fungi the two chains are fused into one protein (Tryptophan synthase, alpha chain, active site). The alpha2beta2 tetramer complex contains pyridoxal-phosphate, glycerol-3-phosphate, Na+ ions. Another key site in tryptophan synthase is the monovalent cation (MVC) site, which is made up of cations like Na+ and K+, along with Cs+ <ref name= "Dierkers"> DOI: 10.1021/bi9008374</ref>. For regulation, TrpA and TrpB cycle between low-activity open conformation and a high-activity closed state. This is done by the binding of an IGP substrate to TrpA which promotes the high activity closed state which activates the TrpB high activity closed state. The states are reset back to low activity open conformation by the production of L-Trp external aldimine <ref name= "Michalska"> PMID: 31316809 </ref>. | ||
| - | + | [Image:LabeledTRPS.JPG]] | |
| - | + | ||
== Function == | == Function == | ||
Revision as of 18:19, 2 May 2022
Tryptophan Synthase
| |||||||||||
References
Miles E. W. (2001). Tryptophan synthase: a multienzyme complex with an intramolecular tunnel. Chemical record (New York, N.Y.), 1(2), 140–151. https://doi.org/10.1002/tcr.4
- ↑ 1.0 1.1 1.2 1.3 Kulik V, Hartmann E, Weyand M, Frey M, Gierl A, Niks D, Dunn MF, Schlichting I. On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes. J Mol Biol. 2005 Sep 23;352(3):608-20. PMID:16120446 doi:10.1016/j.jmb.2005.07.014
- ↑ 2.0 2.1 2.2 2.3 2.4 Michalska K, Gale J, Joachimiak G, Chang C, Hatzos-Skintges C, Nocek B, Johnston SE, Bigelow L, Bajrami B, Jedrzejczak RP, Wellington S, Hung DT, Nag PP, Fisher SL, Endres M, Joachimiak A. Conservation of the structure and function of bacterial tryptophan synthases. IUCrJ. 2019 May 29;6(Pt 4):649-664. doi: 10.1107/S2052252519005955. eCollection, 2019 Jul 1. PMID:31316809 doi:http://dx.doi.org/10.1107/S2052252519005955
- ↑ Dierkers AT, Niks D, Schlichting I, Dunn MF. Tryptophan synthase: structure and function of the monovalent cation site. Biochemistry. 2009 Nov 24;48(46):10997-1010. doi: 10.1021/bi9008374. PMID:19848417 doi:http://dx.doi.org/10.1021/bi9008374
- ↑ 4.0 4.1 Buller AR, van Roye P, Murciano-Calles J, Arnold FH. Tryptophan Synthase Uses an Atypical Mechanism To Achieve Substrate Specificity. Biochemistry. 2016 Dec 27;55(51):7043-7046. doi: 10.1021/acs.biochem.6b01127., Epub 2016 Dec 13. PMID:27935677 doi:http://dx.doi.org/10.1021/acs.biochem.6b01127
- ↑ D'Amico RN, Bosken YK, O'Rourke KF, Murray AM, Admasu W, Chang CA, Boehr DD. Substitution of a Surface-Exposed Residue Involved in an Allosteric Network Enhances Tryptophan Synthase Function in Cells. Front Mol Biosci. 2021 May 26;8:679915. doi: 10.3389/fmolb.2021.679915., eCollection 2021. PMID:34124159 doi:http://dx.doi.org/10.3389/fmolb.2021.679915
- ↑ Abrahams KA, Cox JAG, Futterer K, Rullas J, Ortega-Muro F, Loman NJ, Moynihan PJ, Perez-Herran E, Jimenez E, Esquivias J, Barros D, Ballell L, Alemparte C, Besra GS. Inhibiting mycobacterial tryptophan synthase by targeting the inter-subunit interface. Sci Rep. 2017 Aug 25;7(1):9430. doi: 10.1038/s41598-017-09642-y. PMID:28842600 doi:http://dx.doi.org/10.1038/s41598-017-09642-y
- ↑ doi: https://dx.doi.org/10.1186/1471-2229-8-44</ref==Structuralhighlights==ThisisasamplescenecreatedwithSATto<scenename="/12/3456/Sample/1">color</scene>byGroup,andanothertomake<scenename="/12/3456/Sample/2">atransparentrepresentation</scene>oftheprotein.YoucanmakeyourownscenesonSATstartingfromscratchorloadingandeditingoneofthesesamplescenes.