This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Ceramidase
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
Refs <ref name="Okino">PMID:9603946</ref>, <ref name="Inoue">PMID:19088069</ref> , <ref name="Reverse">PMID:10832092</ref> | Refs <ref name="Okino">PMID:9603946</ref>, <ref name="Inoue">PMID:19088069</ref> , <ref name="Reverse">PMID:10832092</ref> | ||
| - | |||
| - | == Disease == | ||
| - | p<ref name="Okino">PMID:9603946</ref> | ||
| - | == Relevance == | ||
| - | p | ||
== Structural highlights == | == Structural highlights == | ||
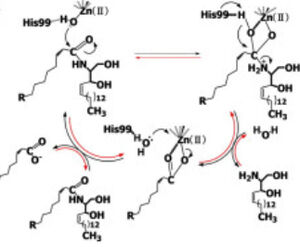
| + | CerN consists of two domains: a catalytic domain near the N-terminal and an immunoglobulin-fold domain near the C-terminal. Three β-sheets, each formed from four β-strands, compose a β-prism fold at the center of the N-terminal domain. Surrounding the β-prism fold are 11 α-helices, forming an α+ β 2-layer sandwich fold. The immunoglobulin C-terminal domain is composed of two β-sheets, containing four β-strands each, forming a β-sandwich fold. Between the N- and C-terminal domains is a magnesium/calcium ion binding site that links together the two domains. His37, Asp579, Asp581, and Thr854 interact with divalent cations within the magnesium/calcium ion binding site. A second metal-binding site containing a zinc ion is located within the N-terminal domain active site, where it is coordinated by His97, His204, Glu411, and a water molecule. | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | + | |
| + | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
<table><tr><td colspan='2'>[[2zxc]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/"bacillus_aeruginosus"_(schroeter_1872)_trevisan_1885 "bacillus aeruginosus" (schroeter 1872) trevisan 1885]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2ZXC OCA]. For a <b>guided tour on the structure components</b> use [http://proteopedia.org/fgij/fg.htm?mol=2ZXC FirstGlance]. <br> | <table><tr><td colspan='2'>[[2zxc]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/"bacillus_aeruginosus"_(schroeter_1872)_trevisan_1885 "bacillus aeruginosus" (schroeter 1872) trevisan 1885]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2ZXC OCA]. For a <b>guided tour on the structure components</b> use [http://proteopedia.org/fgij/fg.htm?mol=2ZXC FirstGlance]. <br> | ||
| Line 22: | Line 19: | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://proteopedia.org/fgij/fg.htm?mol=2zxc FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2zxc OCA], [http://pdbe.org/2zxc PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=2zxc RCSB], [http://www.ebi.ac.uk/pdbsum/2zxc PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=2zxc ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://proteopedia.org/fgij/fg.htm?mol=2zxc FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2zxc OCA], [http://pdbe.org/2zxc PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=2zxc RCSB], [http://www.ebi.ac.uk/pdbsum/2zxc PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=2zxc ProSAT]</span></td></tr> | ||
</table> | </table> | ||
| + | |||
| + | == Disease == | ||
| + | p<ref name="Okino">PMID:9603946</ref> | ||
| + | == Relevance == | ||
| + | p | ||
| + | |||
| + | |||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 18:21, 30 April 2022
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Inoue T, Okino N, Kakuta Y, Hijikata A, Okano H, Goda HM, Tani M, Sueyoshi N, Kambayashi K, Matsumura H, Kai Y, Ito M. Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase. J Biol Chem. 2009 Apr 3;284(14):9566-77. Epub 2008 Dec 16. PMID:19088069 doi:10.1074/jbc.M808232200
- ↑ 2.0 2.1 2.2 2.3 Okino N, Tani M, Imayama S, Ito M. Purification and characterization of a novel ceramidase from Pseudomonas aeruginosa. J Biol Chem. 1998 Jun 5;273(23):14368-73. PMID:9603946
- ↑ 3.0 3.1 Kita K, Okino N, Ito M. Reverse hydrolysis reaction of a recombinant alkaline ceramidase of Pseudomonas aeruginosa. Biochim Biophys Acta. 2000 May 31;1485(2-3):111-20. doi:, 10.1016/s1388-1981(00)00029-9. PMID:10832092 doi:http://dx.doi.org/10.1016/s1388-1981(00)00029-9