Ceramidase
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
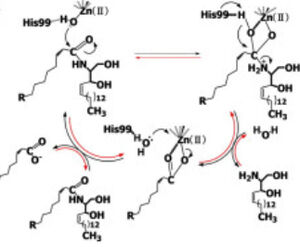
Due to its abundance in eukaryotic cell membranes and role in pathogen defense, pathogens have evolved mechanisms to use host ceramides for their own pathogenesis.<ref name="Seitz">PMID:25720061</ref> Ceramide-rich membrane platforms on human-epithelial cells serve as sites for the '''attachment''' and '''invasion''' of bacterial pathogens such as ''Neisseria gonorrhoeae''.<ref name="Seitz">PMID:25720061</ref> ''Pseudomonas aeruginosa'' is capable of detecting host-derived sphingosine, resulting in activation of ''P. aeruginosa'' sphingosine-responsive genes.<ref name="Annette">PMID:24465209</ref> The products of ''P. aeruginosa'' '''sphingosine-responsive genes''' are used for the detoxification of sphingosine, as well as its production from other host-derived sphingolipids, making ''P. aeruginosa'' '''sphingosine tolerant'''.<ref name="Annette">PMID:24465209</ref> Loss of ''P. aeruginosa'' sphingosine-responsive genes results in the inability of the bacteria to survive in the presence of sphingosine ''in vitro'' or in the murine lung.<ref name="Annette">PMID:24465209</ref> CerN is one of the proteins encoded by ''P. aeruginosa'' sphingosine-responsive genes used for the production of sphingosine from ceramide, with the added ability of functioning as a ceramide synthase.<ref name="Okino">PMID:9603946</ref><ref name="Reverse">PMID:10832092</ref><ref name="Annette">PMID:24465209</ref> It has been hypothesized that the hydrolysis of host-membrane ceramide via CerN facilitates ''P. aeruginosa'' intracellular invasion, similar to other bacterial pathogens.<ref name="Okino">PMID:9603946</ref> CerN is also involved in ''P. aeruginosa'' [https://en.wikipedia.org/wiki/Biofilm biofilm] production, a major virulence trait of the pathogen.<ref name="Theft">PMID:35129518</ref> Sphingosine production via CerN-mediated hydrolysis of host ceramide induces a biofilm formation at sites of infection, biofilm accumulation yields more ceramide hydrolysis, creating a positive-feedback loop for ''P. aeruginosa'' virulence.<ref name="Theft">PMID:35129518</ref> | Due to its abundance in eukaryotic cell membranes and role in pathogen defense, pathogens have evolved mechanisms to use host ceramides for their own pathogenesis.<ref name="Seitz">PMID:25720061</ref> Ceramide-rich membrane platforms on human-epithelial cells serve as sites for the '''attachment''' and '''invasion''' of bacterial pathogens such as ''Neisseria gonorrhoeae''.<ref name="Seitz">PMID:25720061</ref> ''Pseudomonas aeruginosa'' is capable of detecting host-derived sphingosine, resulting in activation of ''P. aeruginosa'' sphingosine-responsive genes.<ref name="Annette">PMID:24465209</ref> The products of ''P. aeruginosa'' '''sphingosine-responsive genes''' are used for the detoxification of sphingosine, as well as its production from other host-derived sphingolipids, making ''P. aeruginosa'' '''sphingosine tolerant'''.<ref name="Annette">PMID:24465209</ref> Loss of ''P. aeruginosa'' sphingosine-responsive genes results in the inability of the bacteria to survive in the presence of sphingosine ''in vitro'' or in the murine lung.<ref name="Annette">PMID:24465209</ref> CerN is one of the proteins encoded by ''P. aeruginosa'' sphingosine-responsive genes used for the production of sphingosine from ceramide, with the added ability of functioning as a ceramide synthase.<ref name="Okino">PMID:9603946</ref><ref name="Reverse">PMID:10832092</ref><ref name="Annette">PMID:24465209</ref> It has been hypothesized that the hydrolysis of host-membrane ceramide via CerN facilitates ''P. aeruginosa'' intracellular invasion, similar to other bacterial pathogens.<ref name="Okino">PMID:9603946</ref> CerN is also involved in ''P. aeruginosa'' [https://en.wikipedia.org/wiki/Biofilm biofilm] production, a major virulence trait of the pathogen.<ref name="Theft">PMID:35129518</ref> Sphingosine production via CerN-mediated hydrolysis of host ceramide induces a biofilm formation at sites of infection, biofilm accumulation yields more ceramide hydrolysis, creating a positive-feedback loop for ''P. aeruginosa'' virulence.<ref name="Theft">PMID:35129518</ref> | ||
| - | |||
| - | |||
== Evolutionary Conservation of Neutral Ceramidases == | == Evolutionary Conservation of Neutral Ceramidases == | ||
| Line 37: | Line 35: | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
| + | |||
| + | ==3D PDB structures of ceramidase== | ||
| + | |||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | {{#tree:id=OrganizedByTopic|openlevels=0| | ||
| + | |||
| + | *Neutral ceramidase | ||
| + | |||
| + | **[[4wgk]] – hCer – human <br /> | ||
| + | **[[2zws]] – PaCer – Pseudomonas aeruginosa <br /> | ||
| + | **[[2zxc]] – PaCer + C2-ceramide <br /> | ||
| + | |||
| + | *Acid ceramidase | ||
| + | |||
| + | **[[5u7z]] – hCer <br /> | ||
| + | **[[6mhm]] – hCer + carmofur <br /> | ||
| + | **[[5u81]] – Cer – mole rat <br /> | ||
| + | **[[5u84]] – Cer – whale<br /> | ||
| + | |||
| + | *Alkaline ceramidase | ||
| + | |||
| + | **[[6g7o]], [[6yxh]] – hCer <br /> | ||
| + | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Revision as of 07:58, 18 January 2023
| |||||||||||