User:Brian Boyle/Sandbox 1
From Proteopedia
| Line 3: | Line 3: | ||
== '''Overview''' == | == '''Overview''' == | ||
| - | |||
'''Peregrin''', also known as Bromodomain and PHD Finger-containing 1 ('''BRPF1''') is a 137 kDa protein that plays a versatile role in epigenetic signaling events. It contains three chromatin reader domains, including a <scene name='91/910741/Apo_bromodomain/1'>bromodomain</scene>, <scene name='91/910741/Apo_pzp/1'>PZP domain</scene> (two PHD fingers separated by a Zinc Knuckle), and '''proline-tryptophan-tryptophan-proline (<scene name='91/910741/Pwwp_unliganded/1'>PWWP</scene>) domain''' (from N to C terminus)<ref name="Yan">PMID:27939640</ref>. Through these three domains, it is capable of recognizing both modified and unmodified histones, as well as non-specifically binding DNA <ref name="Klein">PMID:31711755</ref>,<ref name="Glass1">PMID:24333487</ref>. BRPF1 carries out its function as a component of the MOZ (monocytic leukemic zinc-finger protein) histone acetyltransferase (HAT) complex <ref name="Obi">PMID:33554132</ref>. This complex is involved in the regulation of gene expression, particularly those involved with skeletal development and hematopoiesis <ref>PMID:19254709</ref>,<ref>PMID:27500495</ref>. | '''Peregrin''', also known as Bromodomain and PHD Finger-containing 1 ('''BRPF1''') is a 137 kDa protein that plays a versatile role in epigenetic signaling events. It contains three chromatin reader domains, including a <scene name='91/910741/Apo_bromodomain/1'>bromodomain</scene>, <scene name='91/910741/Apo_pzp/1'>PZP domain</scene> (two PHD fingers separated by a Zinc Knuckle), and '''proline-tryptophan-tryptophan-proline (<scene name='91/910741/Pwwp_unliganded/1'>PWWP</scene>) domain''' (from N to C terminus)<ref name="Yan">PMID:27939640</ref>. Through these three domains, it is capable of recognizing both modified and unmodified histones, as well as non-specifically binding DNA <ref name="Klein">PMID:31711755</ref>,<ref name="Glass1">PMID:24333487</ref>. BRPF1 carries out its function as a component of the MOZ (monocytic leukemic zinc-finger protein) histone acetyltransferase (HAT) complex <ref name="Obi">PMID:33554132</ref>. This complex is involved in the regulation of gene expression, particularly those involved with skeletal development and hematopoiesis <ref>PMID:19254709</ref>,<ref>PMID:27500495</ref>. | ||
| - | |||
== '''PZP Domain''' == | == '''PZP Domain''' == | ||
The PZP domain of BRPF1 has been shown to <scene name='91/910741/Pzp_with_h3/2'>associate with the histone H3 tail</scene>, as well as DNA<ref name="Klein" />. Three residues in the H3 peptide undergo unique interactions with the binding pocket. These are <scene name='91/910741/H3_ala_1/1'>Ala-1</scene>, <scene name='91/910741/Arg_2/1'>Arg-2</scene>, and Thr-3.<ref name="Klein" />. | The PZP domain of BRPF1 has been shown to <scene name='91/910741/Pzp_with_h3/2'>associate with the histone H3 tail</scene>, as well as DNA<ref name="Klein" />. Three residues in the H3 peptide undergo unique interactions with the binding pocket. These are <scene name='91/910741/H3_ala_1/1'>Ala-1</scene>, <scene name='91/910741/Arg_2/1'>Arg-2</scene>, and Thr-3.<ref name="Klein" />. | ||
| Line 16: | Line 14: | ||
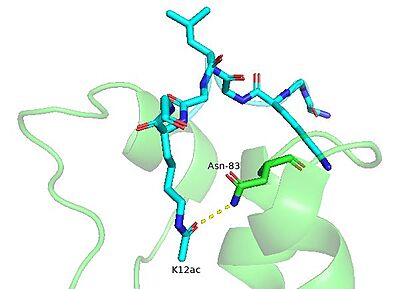
The BRPF1 bromodomain has been shown to recognize and bind to various acetylated lysine marks on the N-terminal tails of histones tails <ref name="Glass1" />. Using isothermal titration calorimetry (ITC) experiments, it was found that the BRPF1 bromodomain preferentially binds to histone H4 acetylated at positions K5 ([[2rs9]]) and K12 ([[4qyd]]) as well as H2A at position K5 ([[4qyl]]) <ref name="Obi" />,<ref name="Glass1" />. Interestingly, the BRPF1 bromodomain has also been shown to bind di-acetylated histone peptides with high affinity, including H4K5acK8ac and H4K5acK12ac <ref name="Obi" />. Acetyllysine recognition is coordinated by a number of residues in the bromodomain's binding pocket. Using NMR chemical shift perturbation experiments, Glass et al. reported several <scene name='91/910741/Nmr_resi_h4_binding/1'>key residues</scene> the undergo conformational changes upon histone H4 ligand binding (I27, L34, E36, V37, N83, and I88)<ref name="Obi" />. Notably, asparagine 83 was among these. The interaction between the amide nitrogen of asparagine with the carbonyl of the acetyllysine group is conserved in all bromodomains and is necessary for binding to occur <ref name ="Obi" />,<ref name ="Lubula_2014" />. | The BRPF1 bromodomain has been shown to recognize and bind to various acetylated lysine marks on the N-terminal tails of histones tails <ref name="Glass1" />. Using isothermal titration calorimetry (ITC) experiments, it was found that the BRPF1 bromodomain preferentially binds to histone H4 acetylated at positions K5 ([[2rs9]]) and K12 ([[4qyd]]) as well as H2A at position K5 ([[4qyl]]) <ref name="Obi" />,<ref name="Glass1" />. Interestingly, the BRPF1 bromodomain has also been shown to bind di-acetylated histone peptides with high affinity, including H4K5acK8ac and H4K5acK12ac <ref name="Obi" />. Acetyllysine recognition is coordinated by a number of residues in the bromodomain's binding pocket. Using NMR chemical shift perturbation experiments, Glass et al. reported several <scene name='91/910741/Nmr_resi_h4_binding/1'>key residues</scene> the undergo conformational changes upon histone H4 ligand binding (I27, L34, E36, V37, N83, and I88)<ref name="Obi" />. Notably, asparagine 83 was among these. The interaction between the amide nitrogen of asparagine with the carbonyl of the acetyllysine group is conserved in all bromodomains and is necessary for binding to occur <ref name ="Obi" />,<ref name ="Lubula_2014" />. | ||
| - | |||
== '''PWWP Domain''' == | == '''PWWP Domain''' == | ||
| + | |||
| + | </StructureSection> | ||
== '''BRPF1 Association with the MOZ HAT Complex''' == | == '''BRPF1 Association with the MOZ HAT Complex''' == | ||
| Line 32: | Line 31: | ||
Mutations within the gene itself have been associated with neurological disorders and widespread reduced histone acetylation <ref name="Yan" />. | Mutations within the gene itself have been associated with neurological disorders and widespread reduced histone acetylation <ref name="Yan" />. | ||
| - | |||
| - | </StructureSection> | ||
== '''Available Structures''' == | == '''Available Structures''' == | ||
Revision as of 19:32, 3 May 2022
| |||||||||||
Contents |
BRPF1 Association with the MOZ HAT Complex
The MOZ Histone Acetyltransferase Complex is a tetramer consisting of MEAF6, ING5, BRPF1 and MOZ or MORF[2]. Within BRPF1, there are two non-chromatin-binding modules surrounding the PZP domain that are responsible for its association with the MOZ HAT Complex. On the N-terminal side of the PZP, lies the MOZ/MORF binding domain[8]. On the other side of the PZP domain, there is a small module involved in binding to ING5 and MEAF6[9]. BRPF1 seems to be required for the formation of the MOZ HAT complex, as it acts as a bridge associating MOZ or MORF with ING5 and MEAF6[9].
Evolutionary Relationships
Bromodomains are categorized into several families based on sequence and structural similarity. The BRPF1 bromodomain belongs to family IV of bromodomains[10].
Links to Human Disease
BRPF1 has been implicated in the progression of several cancers. Chromosomal translocations of the gene encoding MOZ (a subunit in the MOZ HAT complex) have been linked to the development of acute myeloid leukemia [4]. The crucial role of BRPF1 in this complex has made it the subject of many studies in order to understand how this mutation leads to a cancer phenotype. Another study reported an association between upregulation of the BRPF1 gene and poor survival rates in hepatocellular carcinoma patients [11].
Mutations within the gene itself have been associated with neurological disorders and widespread reduced histone acetylation [1].
Available Structures
References
- ↑ 1.0 1.1 Yan K, Rousseau J, Littlejohn RO, Kiss C, Lehman A, Rosenfeld JA, Stumpel CT, Stegmann AP, Robak L, Scaglia F, Nguyen TT, Fu H, Ajeawung NF, Camurri MV, Li L, Gardham A, Panis B, Almannai M, Sacoto MJ, Baskin B, Ruivenkamp C, Xia F, Bi W, Cho MT, Potjer TP, Santen GW, Parker MJ, Canham N, McKinnon M, Potocki L, MacKenzie JJ, Roeder ER, Campeau PM, Yang XJ. Mutations in the Chromatin Regulator Gene BRPF1 Cause Syndromic Intellectual Disability and Deficient Histone Acetylation. Am J Hum Genet. 2017 Jan 5;100(1):91-104. doi: 10.1016/j.ajhg.2016.11.011. Epub, 2016 Dec 8. PMID:27939640 doi:http://dx.doi.org/10.1016/j.ajhg.2016.11.011
- ↑ 2.0 2.1 2.2 2.3 Klein BJ, Cox KL, Jang SM, Cote J, Poirier MG, Kutateladze TG. Molecular Basis for the PZP Domain of BRPF1 Association with Chromatin. Structure. 2019 Nov 6. pii: S0969-2126(19)30355-7. doi:, 10.1016/j.str.2019.10.014. PMID:31711755 doi:http://dx.doi.org/10.1016/j.str.2019.10.014
- ↑ 3.0 3.1 3.2 Poplawski A, Hu K, Lee W, Natesan S, Peng D, Carlson S, Shi X, Balaz S, Markley JL, Glass KC. Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain. J Mol Biol. 2014 Apr 17;426(8):1661-76. doi: 10.1016/j.jmb.2013.12.007. Epub 2013, Dec 12. PMID:24333487 doi:http://dx.doi.org/10.1016/j.jmb.2013.12.007
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Obi JO, Lubula MY, Cornilescu G, Henrickson A, McGuire K, Evans CM, Phillips M, Boyson SP, Demeler B, Markley JL, Glass KC. The BRPF1 bromodomain is a molecular reader of di-acetyllysine. Curr Res Struct Biol. 2020;2:104-115. doi: 10.1016/j.crstbi.2020.05.001. Epub, 2020 May 12. PMID:33554132 doi:http://dx.doi.org/10.1016/j.crstbi.2020.05.001
- ↑ Hibiya K, Katsumoto T, Kondo T, Kitabayashi I, Kudo A. Brpf1, a subunit of the MOZ histone acetyl transferase complex, maintains expression of anterior and posterior Hox genes for proper patterning of craniofacial and caudal skeletons. Dev Biol. 2009 May 15;329(2):176-90. doi: 10.1016/j.ydbio.2009.02.021. Epub 2009 , Feb 27. PMID:19254709 doi:http://dx.doi.org/10.1016/j.ydbio.2009.02.021
- ↑ You L, Li L, Zou J, Yan K, Belle J, Nijnik A, Wang E, Yang XJ. BRPF1 is essential for development of fetal hematopoietic stem cells. J Clin Invest. 2016 Sep 1;126(9):3247-62. doi: 10.1172/JCI80711. Epub 2016 Aug 8. PMID:27500495 doi:http://dx.doi.org/10.1172/JCI80711
- ↑ 7.0 7.1 7.2 Lubula MY, Eckenroth BE, Carlson S, Poplawski A, Chruszcz M, Glass KC. Structural insights into recognition of acetylated histone ligands by the BRPF1 bromodomain. FEBS Lett. 2014 Sep 30. pii: S0014-5793(14)00705-4. doi:, 10.1016/j.febslet.2014.09.028. PMID:25281266 doi:http://dx.doi.org/10.1016/j.febslet.2014.09.028
- ↑ Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, Tan S, Yang XJ, Kutateladze TG, Cote J. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013 Sep 15;27(18):2009-24. doi: 10.1101/gad.223396.113. PMID:24065767 doi:http://dx.doi.org/10.1101/gad.223396.113
- ↑ 9.0 9.1 Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, Champagne N, Cote J, Yang XJ. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol Cell Biol. 2008 Nov;28(22):6828-43. doi: 10.1128/MCB.01297-08. Epub 2008 Sep , 15. PMID:18794358 doi:10.1128/MCB.01297-08
- ↑ Lloyd JT, Glass KC. Biological function and histone recognition of family IV bromodomain-containing proteins. J Cell Physiol. 2018 Mar;233(3):1877-1886. doi: 10.1002/jcp.26010. Epub 2017 Jun , 13. PMID:28500727 doi:http://dx.doi.org/10.1002/jcp.26010
- ↑ Cheng CL, Tsang FH, Wei L, Chen M, Chin DW, Shen J, Law CT, Lee D, Wong CC, Ng IO, Wong CM. Bromodomain-containing protein BRPF1 is a therapeutic target for liver cancer. Commun Biol. 2021 Jul 20;4(1):888. doi: 10.1038/s42003-021-02405-6. PMID:34285329 doi:http://dx.doi.org/10.1038/s42003-021-02405-6