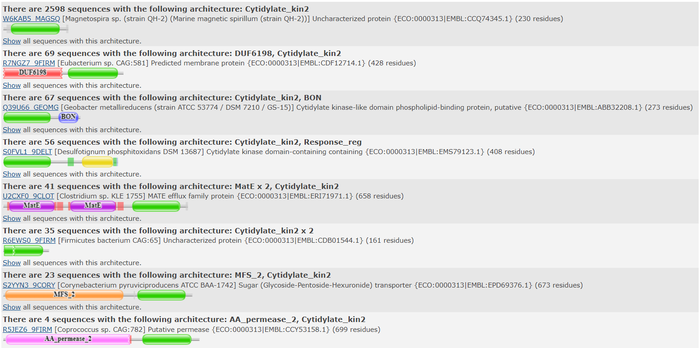
BASIL2022GV3HDT
From Proteopedia
(Difference between revisions)
| Line 96: | Line 96: | ||
===='''Coupled kinase assay'''==== | ===='''Coupled kinase assay'''==== | ||
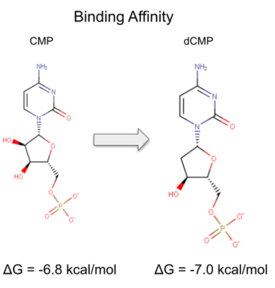
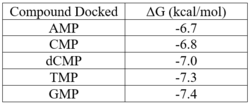
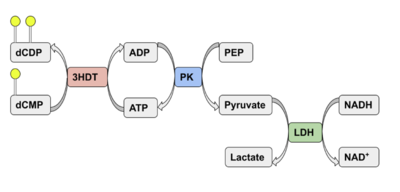
| - | Two rounds of coupled kinase assays were run using 3HDT with ATP and dCMP as substrates. The stock concentration of dCMP was measured to be 109mM, and various concentrations between 5-10mM were used in the assays. The first round of | + | Two rounds of coupled kinase assays were run using 3HDT with ATP and dCMP as substrates. The stock concentration of dCMP was measured to be 109mM, and various concentrations between 5-10mM were used in the assays. The first round of assays (3 assays) used 45.6ng of 3HDT and increasing concentrations of 5.0, 7.5, and 10.0mM dCMP in a well with 100μL total. A concentration of 7.5mM dCMP resulted in the highest specific activity (0.377 U/mg) while increasing the substrate concentration to 10mM had a slightly lower specific activity of 0.348 U/mg. When we repeated the first three kinase assays (5.0, 7.5, 10.0mM dCMP), there were discrepancies in the results indicating a potential experimental error such improper mixing, bubbles being present in the well, or too much time passing between adding substrate to the well and measuring absorbance. Additionally, the protein in the second round of assays was older (original protein but approximately a week had passed from time of over-expression) which could have affected the specific activity in those trials due to the protein starting to expire/decrease in function. |
| Line 108: | Line 108: | ||
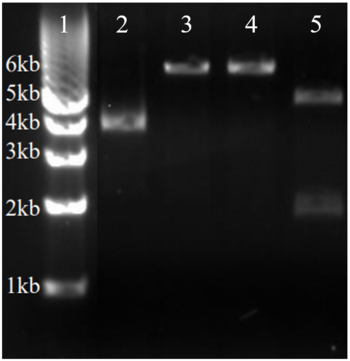
SDS-PAGE results for the purified protein 3HDT. The total weight of this protein is around 25.79 kD. The first lane (left) contains a size standard. The band in the second lane (right) at ~70kD is not the protein of interest (3HDT) but contains a binding metal protein. There is also a faint band around ~26kD, indicating our protein of interest was present. | SDS-PAGE results for the purified protein 3HDT. The total weight of this protein is around 25.79 kD. The first lane (left) contains a size standard. The band in the second lane (right) at ~70kD is not the protein of interest (3HDT) but contains a binding metal protein. There is also a faint band around ~26kD, indicating our protein of interest was present. | ||
| - | [[Image:3hdt sds page.png |225px| center | thumb | Results from running an SDS-PAGE with | + | [[Image:3hdt sds page.png |225px| center | thumb | Results from running an SDS-PAGE with purified 3HDT.]] |
| Line 114: | Line 114: | ||
== Conclusion/Future Experiments == | == Conclusion/Future Experiments == | ||
| - | Our protein was confirmed to be 3HDT using SDS-PAGE and showed activity during coupled kinase assays. While this confirms that 3HDT is a kinase, the true substrate | + | Our protein was confirmed to be 3HDT using SDS-PAGE and showed activity during coupled kinase assays. While this confirms that 3HDT is a kinase, the true substrate was likely not dCMP due to the low specific activity values. Further research should be done with molecules such as TMP and GMP in the future to narrow down potential nucleotide substrates or elucidate other types of compounds to be considered as ligands for 3HDT. Additionally, in the future we would like to repeat the protein purification process to try and get a higher protein concentration than what we achieved as this may have affected our coupled kinase assay results. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 20:44, 5 May 2022
Characterizing Putative Kinase 3HDT
| |||||||||||
References
- ↑ National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2022 April 23].
- ↑ Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
- ↑ Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
- ↑ J. Yu, Y. Zhou, I. Tanaka, M. Yao, Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics, 26(1), 46-52, (2010) [PMID: 19846440]
- ↑ Small-Molecule Library Screening by Docking with PyRx. Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-50.
- ↑ The PyMOL Molecular Graphics System, Version 1.7.4.5 Edu Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Jesse D. Rothfus, Autumn Forrester, Bonnie Hall, Jaime Prilusky