We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
BASIL2022GV3HDT
From Proteopedia
(Difference between revisions)
| Line 54: | Line 54: | ||
== Docking== | == Docking== | ||
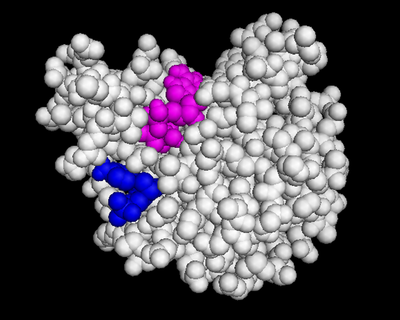
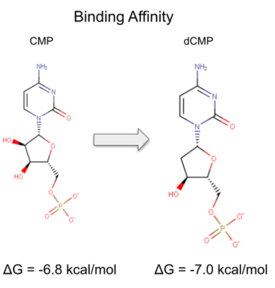
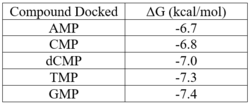
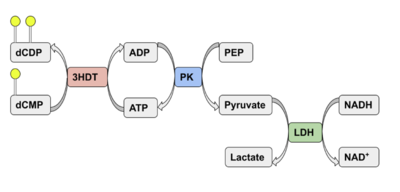
| - | We used POCASA <ref>J. Yu, Y. Zhou, I. Tanaka, M. Yao, Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics, 26(1), 46-52, (2010) [PMID: 19846440]</ref> to determine potential binding pockets within 3HDT before using PyRx<ref>Small-Molecule Library Screening by Docking with PyRx. Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-50.</ref> to dock dCMP into the protein. PyMOL<ref>The PyMOL Molecular Graphics System, Version 1.7.4.5 Edu Schrödinger, LLC.</ref> was then used to visualize the binding pockets with dCMP and other various ligands docked in 3HDT. This <scene name='90/904996/Binding_pockets/1'>binding pocket</scene> (in purple) is a region within 3HDT where the substrate dCMP could bind as indicated by POCASA. This area was also where dCMP bound with the highest affinity in PyRx. The <scene name='90/904996/Hydrophobic_interactions/1'>amino acids</scene> (Gly21, Ser22, Gly23, Val27, Thr 142, Gln149, Arg150, Thr197, Leu200, Thr201) interact within that area to help hold the substrate in place with hydrophobic interactions. Also, there are two residues, <scene name='90/904996/Dcmp_with_3hdt_active_site/1'>Lys146 (purple) and Leu 202 (yellow)</scene> within the active site containing dCMP and cofactor ATP that are important in substrate binding. They interact with the substrate forming hydrogen bonds with the oxygens on the phosphate groups and the 6-membered ring. The program LigPlot <ref>Wallace, A C et al. “LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions.” Protein engineering vol. 8,2 (1995): 127-34. doi:10.1093/protein/8.2.127</ref> was also instrumental in discovering specific interactions between various residues and ligands. However, the PyRx and LigPlot results may be inaccurate due to an issue in the docking process where we were unsuccessful in getting | + | We used POCASA<ref>J. Yu, Y. Zhou, I. Tanaka, M. Yao, Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics, 26(1), 46-52, (2010) [PMID: 19846440]</ref> to determine potential binding pockets within 3HDT before using PyRx<ref>Small-Molecule Library Screening by Docking with PyRx. Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-50.</ref> to dock dCMP into the protein. PyMOL<ref>The PyMOL Molecular Graphics System, Version 1.7.4.5 Edu Schrödinger, LLC.</ref> was then used to visualize the binding pockets with dCMP and other various ligands docked in 3HDT. This <scene name='90/904996/Binding_pockets/1'>binding pocket</scene> (in purple) is a region within 3HDT where the substrate dCMP could bind as indicated by POCASA. This area was also where dCMP bound with the highest affinity in PyRx. The <scene name='90/904996/Hydrophobic_interactions/1'>amino acids</scene> (Gly21, Ser22, Gly23, Val27, Thr 142, Gln149, Arg150, Thr197, Leu200, Thr201) interact within that area to help hold the substrate in place with hydrophobic interactions. Also, there are two residues, <scene name='90/904996/Dcmp_with_3hdt_active_site/1'>Lys146 (purple) and Leu 202 (yellow)</scene> within the active site containing dCMP and cofactor ATP that are important in substrate binding. They interact with the substrate forming hydrogen bonds with the oxygens on the phosphate groups and the 6-membered ring. The program LigPlot<ref>Wallace, A C et al. “LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions.” Protein engineering vol. 8,2 (1995): 127-34. doi:10.1093/protein/8.2.127</ref> was also instrumental in discovering specific interactions between various residues and ligands. However, the PyRx and LigPlot results may be inaccurate due to an issue in the docking process where we were unsuccessful in getting PyRx to recognize where the ATP cofactor was bound before docking our substrate into the protein. |
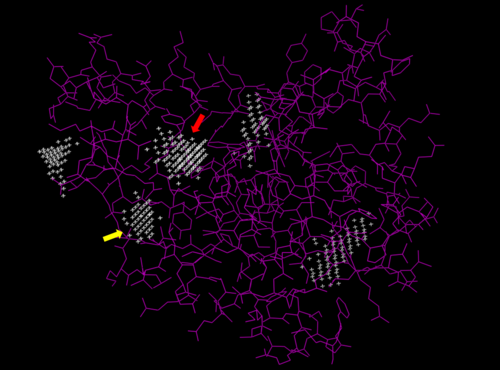
[[Image:POCASA 3hdt image 2.png | 500px| center | thumb| Predicted binding pockets for 3HDT represented by the white stippling.]] | [[Image:POCASA 3hdt image 2.png | 500px| center | thumb| Predicted binding pockets for 3HDT represented by the white stippling.]] | ||
Revision as of 19:26, 7 May 2022
Characterizing Putative Kinase 3HDT and its Potential Substrates
| |||||||||||
References
- ↑ National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2022 April 23].
- ↑ Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
- ↑ Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
- ↑ J. Yu, Y. Zhou, I. Tanaka, M. Yao, Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics, 26(1), 46-52, (2010) [PMID: 19846440]
- ↑ Small-Molecule Library Screening by Docking with PyRx. Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-50.
- ↑ The PyMOL Molecular Graphics System, Version 1.7.4.5 Edu Schrödinger, LLC.
- ↑ Wallace, A C et al. “LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions.” Protein engineering vol. 8,2 (1995): 127-34. doi:10.1093/protein/8.2.127
Proteopedia Page Contributors and Editors (what is this?)
Jesse D. Rothfus, Autumn Forrester, Bonnie Hall, Jaime Prilusky