User:Max Hideki Oliveira Homma/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Function == | == Function == | ||
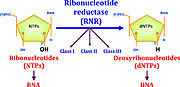
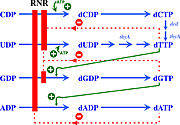
| - | Ribonucleotide reductase (RNR) is an enzyme responsible for converting deoxyribonucleotides (dNTPs) from ribonucleotides (NTPs). It catalyzes this chemical reaction by removing the 2'-hydroxyl group of ribose ring of ribonucleotides. Therefore, this enzyme is also known as ribonucleoside diphosphate reductase (rNDP). In this way, this enzyme promotes the synthesis of precursors for regeneration and construction of DNA. | + | Ribonucleotide reductase (RNR) is an enzyme responsible for converting deoxyribonucleotides (dNTPs) from ribonucleotides (NTPs), as a show in figure 1. It catalyzes this chemical reaction by removing the 2'-hydroxyl group of ribose ring of ribonucleotides. Therefore, this enzyme is also known as ribonucleoside diphosphate reductase (rNDP). In this way, this enzyme promotes the synthesis of precursors for regeneration and construction of DNA. |
[[Image:RNR1.jpg|thumb|'''Figure 1:''' RNRs catalyze the conversion of diphosphated ribonucleotides to their corresponding diphosphate deoxyribonucleotides (Torrents, 2014)]] | [[Image:RNR1.jpg|thumb|'''Figure 1:''' RNRs catalyze the conversion of diphosphated ribonucleotides to their corresponding diphosphate deoxyribonucleotides (Torrents, 2014)]] | ||
| Line 26: | Line 26: | ||
== R1 subunit and active site == | == R1 subunit and active site == | ||
| - | In Ia RNRs the catalytic site is located in the R1 subunit. This subunit is composed of two identical polypeptide chains, thus forming a homodimer. | + | In Ia RNRs the catalytic site is located in the <scene name='91/910665/R1_homodimer/1'>R1 subunit</scene>. This subunit is composed of two identical polypeptide chains, thus forming a homodimer. The secondary structure of each of the <scene name='91/910665/R1_homodimer_rainbow/1'>polypeptide chains</scene> that form the R1 subunit is composed of beta sheets and, mainly, alpha helices. Close to the active site an alpha/beta barrel is formed, which is highly conserved among RNR classes. In addition to the active site, two important allosteric sites are also located in the R1 subunit: the specificity site (S site) and the activity site (A site). |
| - | <scene name='91/910665/R1_homodimer/1'>R1 subunit</scene> | ||
| - | <scene name='91/910665/R1_homodimer_rainbow/1'>R1 monomer</scene> | ||
| - | |||
| - | |||
| - | |||
| - | The catalysis mechanism performed at the RNR active site is represented in the image. | ||
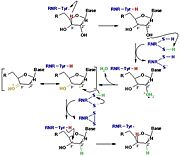
[[Image:RNR3.jpg|thumb|'''Figure 3:''' The reduction of ribonucleotides catalyzed by the RNR involves the participation of cysteines present in the active site]] | [[Image:RNR3.jpg|thumb|'''Figure 3:''' The reduction of ribonucleotides catalyzed by the RNR involves the participation of cysteines present in the active site]] | ||
| - | Initially, the hydrogen bonded to the 3' carbon of the ribose ribonucleotide is transferred to the thiyl radical of Cys439. As a result, the 2'OH radical becomes more accessible for acid hydrolysis and its protonation is catalyzed by Cys225 and Glu441, resulting in the exit of a molecule from the substrate. After that, a proton is transferred from Cys462 to Cys225 which then transfers a hydrogen to the 2' carbon. Then, the transfer of a hydrogen to the 2' carbon of the substrate occurs concomitantly with the formation of a disulfide bridge between the Cys225 and Cys462 of the RNR. Finally, till radical regeneration occurs with the transfer of a hydrogen from Cys439 to the 3' carbon of the substrate, leading to the release of a deoxyribonucleotide. At the end of the catalysis, the disulfide bridge between Cys225 and Cys462 must still be reduced. The electron used in this reduction comes from a redox chain whose initial donor is NADPH. Numerous components participate in this redox chain, including Cys 754 and Cys759 from R1 itself. The reduction of the disulfide bridge between Cys225 and Cys462 would be very difficult to occur if there was a radical very close to the disulfide bridge, since the tendency would be for this radical to be reduced instead of the disulfide bridge. This is a likely explanation for the existence of a second subunit in the RNR, as the R2 subunit harbors a tyrosyl radical that is difficult to reduce. | + | The catalysis mechanism performed at the RNR active site is represented in the figure 3 and depends of <scene name='91/910665/Cys_of_active_site/1'>Cys of active site</scene>.Initially, the hydrogen bonded to the 3' carbon of the ribose ribonucleotide is transferred to the thiyl radical of Cys439. As a result, the 2'OH radical becomes more accessible for acid hydrolysis and its protonation is catalyzed by Cys225 and Glu441, resulting in the exit of a molecule from the substrate. After that, a proton is transferred from Cys462 to Cys225 which then transfers a hydrogen to the 2' carbon. Then, the transfer of a hydrogen to the 2' carbon of the substrate occurs concomitantly with the formation of a <scene name='91/910665/Cys_ball_and_stick/1'>disulfide bridge</scene> between the Cys225 and Cys462 of the RNR. Finally, till radical regeneration occurs with the transfer of a hydrogen from Cys439 to the 3' carbon of the substrate, leading to the release of a deoxyribonucleotide. At the end of the catalysis, the disulfide bridge between Cys225 and Cys462 must still be reduced. The electron used in this reduction comes from a redox chain whose initial donor is NADPH. Numerous components participate in this redox chain, including Cys 754 and Cys759 from R1 itself. The reduction of the disulfide bridge between Cys225 and Cys462 would be very difficult to occur if there was a radical very close to the disulfide bridge, since the tendency would be for this radical to be reduced instead of the disulfide bridge. This is a likely explanation for the existence of a second subunit in the RNR, as the R2 subunit harbors a tyrosyl radical that is difficult to reduce. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== R2 subunit and cofactor == | == R2 subunit and cofactor == | ||
| - | In Ia RNRs, the cofactor binding site is located on the R2 subunit, also known as β. The | + | In Ia RNRs, the cofactor binding site is located on the R2 subunit, also known as β. The R2 subunit may be associated with the α protein in dimeric or oligomeric forms, and there may be one or more β subunits in the same enzyme. |
| - | It contains the essential metal cofactor binding site for the initiation of nucleotide reduction, the ionic transition that occurs with the metallofactor at the | + | It contains the essential metal cofactor binding site for the initiation of nucleotide reduction, the ionic transition that occurs with the metallofactor at the R2 subunit site promotes the formation of the radical that will later be transferred through long-range radical transfer to the alpha subunit, where it will generate the thiol radical at the active site of the enzyme, where the two cysteine loops reduce NTP. |
The R2 protein has a binding site for two Iron atoms on each of its two identical protomers and a nearby potential tyrosyl radical site (Y122 in E.coli). The general structure of the protein is mainly α-helix and each of the di-iron centers are coordinated by aspartate (D84), two histidines (H118 and H241) and three glutamate residues (E115, E204 and E238) that are brought together within a bundle of four helices. This structure is conserved in all subclasses of RNR I, with very few exceptions. | The R2 protein has a binding site for two Iron atoms on each of its two identical protomers and a nearby potential tyrosyl radical site (Y122 in E.coli). The general structure of the protein is mainly α-helix and each of the di-iron centers are coordinated by aspartate (D84), two histidines (H118 and H241) and three glutamate residues (E115, E204 and E238) that are brought together within a bundle of four helices. This structure is conserved in all subclasses of RNR I, with very few exceptions. | ||
| Line 53: | Line 43: | ||
| - | All | + | All R2 subunits belong to the ferritin structural superfamily, with a core composed of a package of four central helices that connect to assemble the active oxidant. In class Ia, this core unit is symmetrical, with two α-helix pairs arranged in a head-to-tail direction to sink the Metallocofactor, FeIII-O-FeIII in this class, into the β structure and protect it from the solvent. Each of the helices in the pair confer a linker region with the cofactor. In addition to cofactor protection, the helical structure also provides second-sphere side chains that contribute to cofactor assembly and reactivity. |
<scene name='91/910665/R2_subunit/1'>R2 subunit homodimer</scene> | <scene name='91/910665/R2_subunit/1'>R2 subunit homodimer</scene> | ||
Revision as of 19:37, 16 June 2022
</math>==Your Heading Here (maybe something like 'Structure')==
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644