Journal:MicroPubl Biol:000606
From Proteopedia
(Difference between revisions)

| Line 14: | Line 14: | ||
These conserved motifs mediate the binding of RNA and/or the binding and hydrolysis of ATP. Interestingly, Utp25 has significant sequence changes in most of these motifs. Mutational loss of the remaining conserved sequence motifs 1a and partial <scene name='92/920755/Cv2/15'>motif VI</scene> resulted in no change in growth <ref name='CharetteBaserga2010'/>. | These conserved motifs mediate the binding of RNA and/or the binding and hydrolysis of ATP. Interestingly, Utp25 has significant sequence changes in most of these motifs. Mutational loss of the remaining conserved sequence motifs 1a and partial <scene name='92/920755/Cv2/15'>motif VI</scene> resulted in no change in growth <ref name='CharetteBaserga2010'/>. | ||
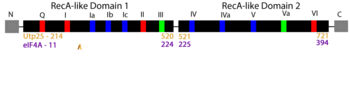
| - | Using the AlphaFold predicted yeast Utp25 structure as a query, we used Dali to search for proteins with a similar structure. Our top hits were to other DEAD-box helicases including the prototypical RNA helicase eIF4A. We then used Chimera to structurally align Utp25 and | + | Using the AlphaFold predicted yeast Utp25 structure as a query, we used Dali to search for proteins with a similar structure. Our top hits were to other DEAD-box helicases including the prototypical RNA helicase eIF4A. We then used Chimera to structurally align Utp25 and eIF4A. By independently aligning the structures of <scene name='92/920755/Cv/9'>domain 1</scene> from Utp25 ([https://alphafold.ebi.ac.uk/entry/P40498 P40498]; gold) and eIF4A ([[1fuu]]; medium violet red) and similarly of <scene name='92/920755/Cv4/2'>domain 2</scene>, we show that Utp25 globally adopts a structure that is very similar to that of DEAD-box RNA helicases. |
'''DEAD-box motifs:''' | '''DEAD-box motifs:''' | ||
'''Domain 1''' | '''Domain 1''' | ||
| - | *<scene name='92/920755/Cv2/4'>Motif Q - GFEEPSAIQ</scene> (red | + | *<scene name='92/920755/Cv2/4'>Motif Q - GFEEPSAIQ</scene> (red eIF4A/pink Utp25). |
| - | *<scene name='92/920755/Cv2/3'>Motif I - AQSGTGKT</scene> (red | + | *<scene name='92/920755/Cv2/3'>Motif I - AQSGTGKT</scene> (red eIF4A/pink Utp25). |
| - | *<scene name='92/920755/Cv2/5'>Motif Ia - PTRELA</scene> (blue | + | *<scene name='92/920755/Cv2/5'>Motif Ia - PTRELA</scene> (blue eIF4A/deepskyblue Utp25). |
| - | *<scene name='92/920755/Cv2/6'>Motif Ib - GG</scene> (blue | + | *<scene name='92/920755/Cv2/6'>Motif Ib - GG</scene> (blue eIF4A/deepskyblue Utp25). |
| - | *<scene name='92/920755/Cv2/7'>Motif Ic - TPGRV</scene> (blue | + | *<scene name='92/920755/Cv2/7'>Motif Ic - TPGRV</scene> (blue eIF4A/deepskyblue Utp25). |
| - | *<scene name='92/920755/Cv2/8'>Motif II - DEAD</scene> (red | + | *<scene name='92/920755/Cv2/8'>Motif II - DEAD</scene> (red eIF4A/pink Utp25). |
| - | *<scene name='92/920755/Cv2/9'>Motif III - SAT</scene> (green | + | *<scene name='92/920755/Cv2/9'>Motif III - SAT</scene> (green eIF4A/palegreen Utp25). |
'''Domain 2''' | '''Domain 2''' | ||
| - | *<scene name='92/920755/Cv2/10'>Motif IV - VIFCNTRR</scene> (blue | + | *<scene name='92/920755/Cv2/10'>Motif IV - VIFCNTRR</scene> (blue eIF4A/deepskyblue Utp25). |
| - | *<scene name='92/920755/Cv2/11'>Motif IVa - AIYSDLPQQERDTIMKEFR</scene> (blue | + | *<scene name='92/920755/Cv2/11'>Motif IVa - AIYSDLPQQERDTIMKEFR</scene> (blue eIF4A/deepskyblue Utp25). |
| - | *<scene name='92/920755/Cv2/14'>Motif V - LISTDLL</scene> (blue | + | *<scene name='92/920755/Cv2/14'>Motif V - LISTDLL</scene> (blue eIF4A/deepskyblue Utp25). |
| - | *<scene name='92/920755/Cv2/13'>Motif Va - ARGIDVQQVSLVINYD</scene> (green | + | *<scene name='92/920755/Cv2/13'>Motif Va - ARGIDVQQVSLVINYD</scene> (green eIF4A/palegreen Utp25). |
| - | *<scene name='92/920755/Cv2/15'>Motif VI - HRIGRGGR</scene> (red | + | *<scene name='92/920755/Cv2/15'>Motif VI - HRIGRGGR</scene> (red eIF4A/pink Utp25). |
| - | The colour of the DEAD-box motifs is derived from the static image (see above) with Utp25 in the lighter colour (such as pink) and | + | The colour of the DEAD-box motifs is derived from the static image (see above) with Utp25 in the lighter colour (such as pink) and eIF4A in the darker colour (such as red). Secondary structures flanking the DEAD-box motifs are coloured gold (Utp25) and medium violet red (eIF4A). DEAD-box motif sequences (based on <ref name='PutnamJankowsky' />) are from the yeast eIF4A. |
Examination of the helicase motifs similarly shows that they are superimposable, except for <scene name='92/920755/Cv2/6'>motif Ib</scene>. This local structural similarity has been maintained despite the sequence divergence of the helicase motifs in Utp25 (except for <scene name='92/920755/Cv2/5'>motif Ia</scene> and partial <scene name='92/920755/Cv2/15'>motif VI</scene> that have maintained sequence conservation). Thus, we propose that Utp25 is a pseudohelicase based on it being an essential protein that adopts a helicase structure - both globally and locally - while having lost the catalytic sequence motifs (with remaining motifs being dispensable). | Examination of the helicase motifs similarly shows that they are superimposable, except for <scene name='92/920755/Cv2/6'>motif Ib</scene>. This local structural similarity has been maintained despite the sequence divergence of the helicase motifs in Utp25 (except for <scene name='92/920755/Cv2/5'>motif Ia</scene> and partial <scene name='92/920755/Cv2/15'>motif VI</scene> that have maintained sequence conservation). Thus, we propose that Utp25 is a pseudohelicase based on it being an essential protein that adopts a helicase structure - both globally and locally - while having lost the catalytic sequence motifs (with remaining motifs being dispensable). | ||
Revision as of 01:55, 19 September 2022
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.