4eb6
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
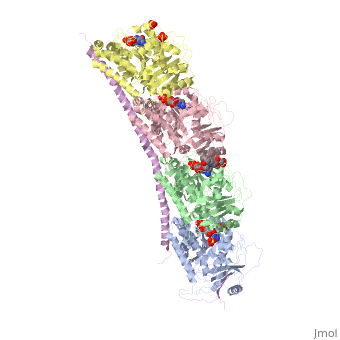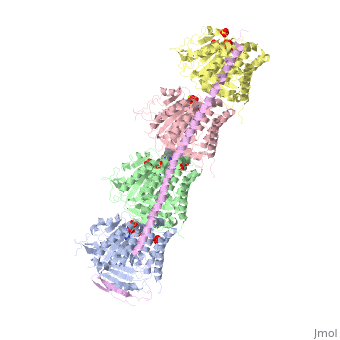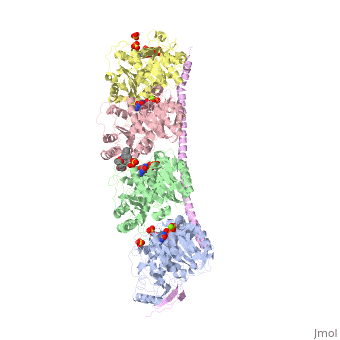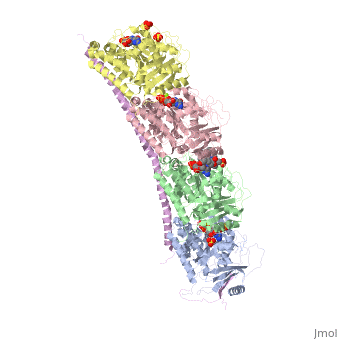
<table><tr><td colspan='2'>[[4eb6]] is a 5 chain structure with sequence from [https://en.wikipedia.org/wiki/Ovis_aries Ovis aries] and [https://en.wikipedia.org/wiki/Rattus_norvegicus Rattus norvegicus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4EB6 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4EB6 FirstGlance]. <br> | <table><tr><td colspan='2'>[[4eb6]] is a 5 chain structure with sequence from [https://en.wikipedia.org/wiki/Ovis_aries Ovis aries] and [https://en.wikipedia.org/wiki/Rattus_norvegicus Rattus norvegicus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4EB6 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4EB6 FirstGlance]. <br> | ||
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GDP:GUANOSINE-5-DIPHOSPHATE'>GDP</scene>, <scene name='pdbligand=GTP:GUANOSINE-5-TRIPHOSPHATE'>GTP</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=VLB:(2ALPHA,2BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE'>VLB</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.47Å</td></tr> |
| + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GDP:GUANOSINE-5-DIPHOSPHATE'>GDP</scene>, <scene name='pdbligand=GTP:GUANOSINE-5-TRIPHOSPHATE'>GTP</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=VLB:(2ALPHA,2BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE'>VLB</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4eb6 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4eb6 OCA], [https://pdbe.org/4eb6 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4eb6 RCSB], [https://www.ebi.ac.uk/pdbsum/4eb6 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4eb6 ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4eb6 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4eb6 OCA], [https://pdbe.org/4eb6 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4eb6 RCSB], [https://www.ebi.ac.uk/pdbsum/4eb6 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4eb6 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/D0VWZ0_SHEEP D0VWZ0_SHEEP] Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).[RuleBase:RU003505][SAAS:SAAS023123_004_019801] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==See Also== | ==See Also== | ||
*[[Stathmin-4 3D structures|Stathmin-4 3D structures]] | *[[Stathmin-4 3D structures|Stathmin-4 3D structures]] | ||
*[[Tubulin 3D Structures|Tubulin 3D Structures]] | *[[Tubulin 3D Structures|Tubulin 3D Structures]] | ||
| - | == References == | ||
| - | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
Current revision
Tubulin-Vinblastine: Stathmin-like complex
| |||||||||||