Sandbox Reserved 1732
From Proteopedia
| Line 2: | Line 2: | ||
==Insulin Receptor Protein== | ==Insulin Receptor Protein== | ||
<Structure load='6SOF' size='400' frame='true' align='right' caption='Insulin Receptor' scene='Insert optional scene name here' /> | <Structure load='6SOF' size='400' frame='true' align='right' caption='Insulin Receptor' scene='Insert optional scene name here' /> | ||
| - | |||
<scene name='91/919041/Insulin_receptor/1'>Text To Be Displayed</scene> | <scene name='91/919041/Insulin_receptor/1'>Text To Be Displayed</scene> | ||
| + | |||
| + | ==Insulin Receptor Structure == | ||
| + | <StructureSection load='6SOF' size='350' side='right' caption='Alpha Chain of Insulin Receptor' scene=''> | ||
| + | |||
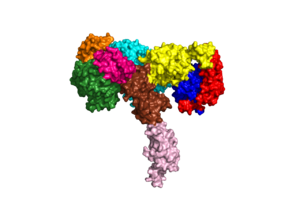
| + | An insulin receptor is a dimer of heterodimers. The dimers are noncovalent, but the insulin receptors are covalently maintained as functional dimers by disulfide bonds. An insulin receptor is comprised of 2 α-chains, and 2 β-chains. The α-chain and an estimated 190 residues of the β-chain are located on the extracellular side of the plasma membrane. The rest of the beta-chain consists of a single transmembrane helix, the juxtamembrane domain, and the intracellular tyrosine kinase domain. | ||
| + | The alpha subunits are the site for insulin binding. Each subunit is comprised of 2 Leucine rich domains (L1 and L2), a Cysteine rich domain (CR) and an α-chain C-terminal helix (α-CT). The two subunits are held together by a disulfide bond between the cysteine rich domains. | ||
| + | |||
| + | Alpha Chain | ||
| + | |||
| + | </StructureSection> | ||
This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
Revision as of 00:40, 15 November 2022
| This Sandbox is Reserved from August 30, 2022 through May 31, 2023 for use in the course Biochemistry I taught by Kimberly Lane at the Radford University, Radford, VA, USA. This reservation includes Sandbox Reserved 1730 through Sandbox Reserved 1749. |
To get started:
More help: Help:Editing |
Contents |
Insulin Receptor Protein
|
Insulin Receptor Structure
| |||||||||||
This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
What is an Insulin Receptor?
An insulin receptor is a large protein that binds to insulin and passes its message into the cell. When insulin binds to the receptor, it is activated and triggers a series of chemical reactions within the cell. These reactions cause an uptake in glucose and other metabolic and growth-related functions. The insulin receptor binds to insulin that is produced in our bodies as well as FDA approved designer insulin which allows people with diabetes to maintain the blood sugar levels necessary. Along with controlling homeostasis, insulin receptors also play a crucial role in regulating lipid, protein, and carbohydrate metabolism, as well as modulating brain neurotransmitter levels [3].
Function
Insulin receptors are regions outside of a cell that allow the cell to bind with the insulin in the bloodstream. The main physiological role of insulin receptors is metabolic regulation.
In order for the receptors to properly interact with Insulin, Insulin must undergo a conformational change. This conformational change is when the C- terminal of the B chain must disengage from the hormone's core. Now the Insulin can properly bind to the appropriate receptor.
Insulin
In order to fully comprehend insulin receptors, it is important to understand what insulin is. Insulin is a hormone that allows our bodies to absorb glucose that is found in our bloodstream. The main source of insulin in the body is from the pancreas, which consists of islets. Islets are what produce and determine the amount of insulin based on the body's blood glucose levels. The higher the glucose levels, the more insulin is produced and released in order to bring blood sugar levels back to an equilibrium.
Disease
Relevance
Structural highlights
An insulin receptor is a dimer of heterodimers. The dimers are noncovalent, but the insulin receptors are covalently maintained as functional dimers by disulfide bonds. An insulin receptor is comprised of 2 α-chains, and 2 β-chains. The α-chain and an estimated 190 residues of the β-chain are located on the extracellular side of the plasma membrane. The rest of the beta-chain consists of a single transmembrane helix, the juxtamembrane domain, and the intracellular tyrosine kinase domain. The alpha subunits are the site for insulin binding. Each subunit is comprised of 2 Leucine rich domains (L1 and L2), a Cysteine rich domain (CR) and an α-chain C-terminal helix (α-CT). The two subunits are held together by a disulfide bond between the cysteine rich domains.
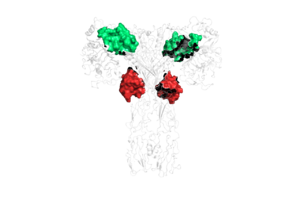
Due to the heterodimeric nature of the receptor, there are two types of insulin binding sites that are split into pairs in the alpha subunits: sites 1 and 1' and sites 2 and 2', for a total of 4 binding sites of insulin. Binding sites 1 and 1' have a greater surface area and are more easily accessible for the insulin to bond, resulting in a higher affinity for insulin binding. Binding sites 2 and 2' have less surface area and are located on the back of the beta sheet so their binding sites do not get filled as quickly.The beta subunits consist of Fibronectin domain III-2 (FnIII-2) and Fibronectin domain III-3 (FnIII-3).
There are two main conformations of full-length insulin receptors, U-shaped and T-shaped. The U-shaped conformation exists when there is an absence of insulin on the protein, whereas the T-shaped and II-shaped conformations exist in the presence of insulin. The U-shaped conformation is also known as the auto-inhibited state. In this state, the L1 domain contacts the L1’ and L2’ domains of the partnering protein. When insulin binds to the receptor protein, it causes a conformational change from the U-shape to the T-shape conformation.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
1. PDB101: Molecule of the month: Insulin receptor https://pdb101.rcsb.org/motm/182 (accessed Oct 21, 2022).