We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Cell division protein
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== General Function == | == General Function == | ||
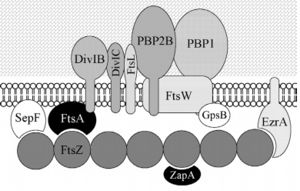
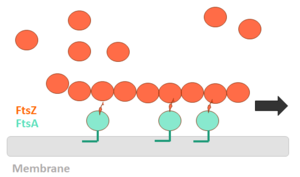
| - | FtsZ (Filamentation temperature sensitive Z) is the main coordinator of septum formation and the most widely conserved division protein, being present in essentially all bacterial genomes that have been sequenced to date. In eubacteria, the FtsZ gene is usually found in the dcw gene cluster, a DNA region containing division and cell-wall synthesis gene. This protein is extremely important to binary fission - more specifically, formation of the Z ring - in rod-shaped bacteria entails the formation of a transverse septum that divides a progenitor cell into two equal-sized daughter cells. Septum formation is faithfully coordinated with chromosome replication and segregation and the spatial control, on the other hand, is evident in the placement of the newly formed septum at precisely the middle of the progenitor cell, which ensures that the two daughter cells generated are morphologically and genetically equivalent. It means that despite the apparent simplicity, cell division in bacteria is subject to tight spatiotemporal control. | + | '''FtsZ''' (Filamentation temperature sensitive Z) is the main coordinator of septum formation and the most widely conserved division protein, being present in essentially all bacterial genomes that have been sequenced to date<ref>PMID:32692252</ref>. In eubacteria, the FtsZ gene is usually found in the dcw gene cluster, a DNA region containing division and cell-wall synthesis gene. This protein is extremely important to binary fission - more specifically, formation of the Z ring - in rod-shaped bacteria entails the formation of a transverse septum that divides a progenitor cell into two equal-sized daughter cells. Septum formation is faithfully coordinated with chromosome replication and segregation and the spatial control, on the other hand, is evident in the placement of the newly formed septum at precisely the middle of the progenitor cell, which ensures that the two daughter cells generated are morphologically and genetically equivalent. It means that despite the apparent simplicity, cell division in bacteria is subject to tight spatiotemporal control. |
== Structure and Biochemistry of FtsZ == | == Structure and Biochemistry of FtsZ == | ||
Revision as of 07:37, 17 November 2022
| |||||||||||
References
[2] [3] [4] [5] [6] [7] [8] [9]
- ↑ Silber N, Matos de Opitz CL, Mayer C, Sass P. Cell division protein FtsZ: from structure and mechanism to antibiotic target. Future Microbiol. 2020 Jun;15:801-831. doi: 10.2217/fmb-2019-0348. Epub 2020 Jul , 21. PMID:32692252 doi:http://dx.doi.org/10.2217/fmb-2019-0348
- ↑ Bisson-Filho AW, Discola KF, Castellen P, Blasios V, Martins A, Sforca ML, Garcia W, Zeri AC, Erickson HP, Dessen A, Gueiros-Filho FJ. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc Natl Acad Sci U S A. 2015 Apr 6. pii: 201414242. PMID:25848052 doi:http://dx.doi.org/10.1073/pnas.1414242112
- ↑ Wang, X. & Lutkenhaus, J. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria.Mol. Microbiol. 21, 313–319 (1996). Gueiros-Filho, F. J. & Losick, R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 (2002).
- ↑ Wang, X., Huang, J., Mukherjee, A., Cao, C., and Lutkenhaus, J. (1997). Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179, 5551–5559.
- ↑ Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol. 2004 Jan;58(1):19-29. doi: 10.1007/s00239-003-2523-5. PMID:14743312 doi:http://dx.doi.org/10.1007/s00239-003-2523-5
- ↑ Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife. 2014 Dec 9;3:e04601. doi: 10.7554/eLife.04601. PMID:25490152 doi:http://dx.doi.org/10.7554/eLife.04601
- ↑ Huecas, S. et al. Energetics and geometry of FtsZ polymers: nucleated self-assembly of single protofilaments. Biophys. J. 94, 1796–1806 (2008).
- ↑ Lan, G. et al. (2009) Condensation of FtsZ filaments can drive bacterial cell division. Proc. Natl. Acad. Sci. U. S. A. 106, 121–126
- ↑ FILHO, Frederico Gueiros. Cell Division. In: GRAUMANN, Peter L. et al. Bacillus: Cellular and Molecular Biology. Germany: Caister Academic Press, 2017.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Jonathan Cardoso C. Vieira, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky