We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1761
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
== Important amino acids== | == Important amino acids== | ||
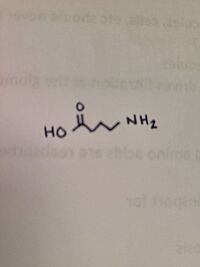
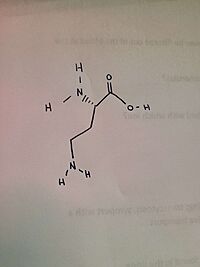
The ligand of ''h''OAT is Pyridoxal-5'-Phosphate <scene name='93/934005/Plp/1'>(PLP)</scene>. The role of the catalytic amino acids in an enzyme is to bind to a substrate, changing the structure, causing bonds to break and new bonds to be formed. When there is a difficult reaction, the triad of amino acids works in tandem to facilitate the reaction. A <scene name='93/934005/Glu235/1'>Glu 235</scene>-<scene name='93/934005/Arg413/1'>Arg 413</scene> <scene name='93/934005/Arg_413_glu_235/1'>salt bridge</scene> was found on AVA, but not on GABA. In regards to GABA, the salt bridge was disrupted in 30% of the monomers. | The ligand of ''h''OAT is Pyridoxal-5'-Phosphate <scene name='93/934005/Plp/1'>(PLP)</scene>. The role of the catalytic amino acids in an enzyme is to bind to a substrate, changing the structure, causing bonds to break and new bonds to be formed. When there is a difficult reaction, the triad of amino acids works in tandem to facilitate the reaction. A <scene name='93/934005/Glu235/1'>Glu 235</scene>-<scene name='93/934005/Arg413/1'>Arg 413</scene> <scene name='93/934005/Arg_413_glu_235/1'>salt bridge</scene> was found on AVA, but not on GABA. In regards to GABA, the salt bridge was disrupted in 30% of the monomers. | ||
| - | The crystal soaking experiments created an opportunity to decipher the reaction intermediates of the structures. While GABA linked to PLP covalently, PLP and catalytic Lys 292 were linked by AVA. | + | The crystal soaking experiments created an opportunity to decipher the reaction intermediates of the structures. While GABA linked to PLP covalently, PLP and catalytic <scene name='93/934005/Lys_292/1'>Lys 292</scene> were linked by AVA. |
== Structural highlights == | == Structural highlights == | ||
Current revision
| This Sandbox is Reserved from November 4, 2022 through January 1, 2023 for use in the course CHEM 351 Biochemistry taught by Bonnie Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1755 through Sandbox Reserved 1764. |
To get started:
More help: Help:Editing |
Human ornithine aminotransferase (hOAT)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ https://doi.org/10.1016/j.jbc.2022.101969
Butrin, A., Butrin, A., Wawrzak, Z., Moran, G. R., & Liu, D. (2022). Determination of the ph dependence, substrate specificity, and turnovers of alternative substrates for human ornithine aminotransferase. Journal of Biological Chemistry, 298(6), 101969. https://doi.org/10.1016/j.jbc.2022.101969