|
General Information
 Organization of the biological protein NKX2.5. The structures analyzed only represent the homeodomain region. The transcription factor, NKX2.5 is one of many proteins classified as a homeodomain, and functions to regulate structural development in eukaryotes. These proteins share a characteristic evolutionarily conserved fold containing . [1]. DNA-binding is mediated through the insertion of the C-terminal side (alpha-3) into the major groove, allowing for base-reside interactions. This allows homeodomains to locate and bind specific DNA sequences, leading to transcriptional activation or repression [2]. The homeodomain of NKX2.5 is flanked by both a N and C-terminal regulatory domain. This puts the biological protein at 324 residues with the homeodomain consisting of residues 138-197 [3] Research into the structure and function of NKX2.5 has mainly been focused on the DNA-binding homeodomain, as mutations in this region have been linked to specific diseases [4]. This page will focus on the specific interactions of the homeodomain of NKX2.5 with DNA, and how these interactions relate to one of the transcription factor's primary function - heart development.
Clinical Relevance
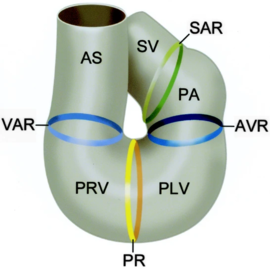 The sinus venosus (SV), the sinoatrial ring (SAR), the primitive atrium (PA), atrioventricular ring (AVR), primitive left ventricle (PLV), primary ring (PR), primitive right ventricle (PRV), ventriculoarterial ring (VAR), and the aortic sac (AS) [5]. In the case of NKX2.5, the protein works in conjunction with multiple other transcription factors during cardiogenesis [6],[7]. Recently, research has been focused on NKX2.5 as mutations in the DNA binding residues, and structural support residues of the protein have been linked to congenital heart disease. Specifically, NKX2.5 mutations have been linked to etiologies involving both atrial and septal defects, deficient atrioventricular node conduction, and more complex mutations such as Tetralogy of Fallot and Hypoplastic Left Heart Syndrome [8],[4],[9]. These phenotypes are speculated to arise as a result of decreased DNA-binding of NKX2.5 during cardiogenesis. Thi [10]. Specifically, issues take root during cardiac loop formation, which takes place approximately 24 days after embryogenesis. At this time, the single vessel present in the embryo loops back upon itself, allowing the vessel to come in contact with itself. This leads to the formation of a four-chambered, primitive heart with the great vessels (shown in the figure from Gittenberger-de Groot et. al)[5]. Throughout this point in development, NKX2.5 along with other transcription factors are essential in facilitating initial loop formation. Therefore, mutations that affect NKX2.5 function have been tied to the defects mentioned above [11].
Additionally, studies have shown that NKX2.5 continues to function after development by maintaining conductivity through the heart's natural pace-maker system. Although the process by which this occurs is slightly less well known, it does suggest NKX2.5 is important throughout life [12]. Moreover, NKX2.5 continues to function as a transcription factor for other cardiac-related peptides, for example atrial natriuretic factors [13].
Structural highlights
Protein / DNA Interactions
DNA-binding is mediated by residues of probing into the major groove. In the structure presented, NKX2.5 is binding to a known target: the ANF-242 sequence downstream of the atrial natriuretic factor peptide promoter [4]. The ANF-242 site contains two NK family binding motifs, , separated by a five nucelotide gap [14]. NKX2.5 interacts to this motif with residues positioned on alpha helix 3. Thre residues, interact with DNA backbone at the NK binding motif. Interactions to bases in the major groove is mediated by two residues: . In addition to these two major grove interactions, there are three additional reidues that interact with bases in the NK motif. These bases are . These residues are of particular importance as mutations in this region has been directly linked to pathologies discussed above [4],[15].
Protein / Protein Interactions
The global structure of NKX2.5 is maintained by both hydrophobic and hydrophilic interactions between the three alpha-helices. This is clear when observing the structure of NKX2.5 where . Besides these tertiary interactions present in NKX2.5, the protein is known to associate with other transcription factors and regulatory proteins. For example, GATA factors, and the Hand1 transcription factor (both of which are important in cardiogenesis) are known to interact with NKX2.5 [16],[17]. Unfortunately, structures are not available for these complexes. Structures have been elucidated for the interaction of (T-box5)[18]. This interaction appears is thought to be mediated : Lys 158 of NKX2.5 along with Asp 140 and Pro 142 from TBX5. A complex formed between (myocyte enhancer factor 2) has also been elucidated [19].
Evolution
The homeodomain of NKX2.5 is highly conserved across the animal kingdom [1]. This can easily be seen in a sequence alignment containing NKX2.5 isoforms across a variety of species. A selection of the Clustal Omega sequence alignment [20] shown below outlines the conservation of the homeodomain of NKX2.5.
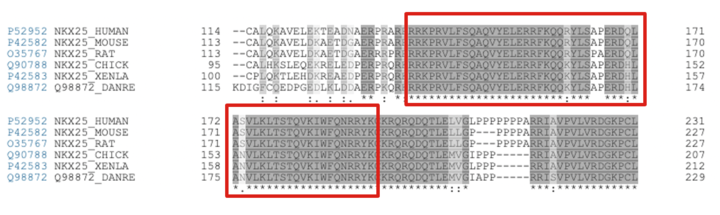 Sequence alignment of NKX2.5 isoforms from multiple species. Red boxes indicate the continuous homeodomain region from residue 137-194. It is important to note that the residues involved in the recognition of the NKX2.5 motif are fully conserved across all isoforms. The cross-species similarities in NKX2.5 isozymes is of special importance in relation to research. For example, Danio rerio (Zebrafish) have been used as models to study heart development during gestation [21], [22]. This is made possible by the fact that the Danio rerio and Homo sapien NKX2.5 share sequence similarities.
References
- ↑ 1.0 1.1 Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487-526. doi: 10.1146/annurev.bi.63.070194.002415. PMID:7979246 doi:http://dx.doi.org/10.1146/annurev.bi.63.070194.002415
- ↑ Burglin TR, Affolter M. Homeodomain proteins: an update. Chromosoma. 2016 Jun;125(3):497-521. doi: 10.1007/s00412-015-0543-8. Epub 2015, Oct 13. PMID:26464018 doi:http://dx.doi.org/10.1007/s00412-015-0543-8
- ↑ Pradhan L, Genis C, Scone P, Weinberg EO, Kasahara H, Nam HJ. Crystal structure of the human NKX2.5 homeodomain in complex with DNA target. Biochemistry. 2012 Aug 14;51(32):6312-9. Epub 2012 Aug 3. PMID:22849347 doi:http://dx.doi.org/10.1021/bi300849c
- ↑ 4.0 4.1 4.2 4.3 Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998 Jul 3;281(5373):108-11. PMID:9651244
- ↑ 5.0 5.1 Gittenberger-de Groot AC, Bartelings MM, Deruiter MC, Poelmann RE. Basics of cardiac development for the understanding of congenital heart malformations. Pediatr Res. 2005 Feb;57(2):169-76. doi: 10.1203/01.PDR.0000148710.69159.61. Epub, 2004 Dec 20. PMID:15611355 doi:http://dx.doi.org/10.1203/01.PDR.0000148710.69159.61
- ↑ Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006 Sep 29;313(5795):1922-7. doi: 10.1126/science.1132292. PMID:17008524 doi:http://dx.doi.org/10.1126/science.1132292
- ↑ Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacol Ther. 2005 Aug;107(2):252-68. doi: 10.1016/j.pharmthera.2005.03.005. PMID:15925411 doi:http://dx.doi.org/10.1016/j.pharmthera.2005.03.005
- ↑ Toko H, Zhu W, Takimoto E, Shiojima I, Hiroi Y, Zou Y, Oka T, Akazawa H, Mizukami M, Sakamoto M, Terasaki F, Kitaura Y, Takano H, Nagai T, Nagai R, Komuro I. Csx/Nkx2-5 is required for homeostasis and survival of cardiac myocytes in the adult heart. J Biol Chem. 2002 Jul 5;277(27):24735-43. doi: 10.1074/jbc.M107669200. Epub 2002 , Mar 11. PMID:11889119 doi:http://dx.doi.org/10.1074/jbc.M107669200
- ↑ McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003 Nov 5;42(9):1650-5. doi: 10.1016/j.jacc.2003.05.004. PMID:14607454 doi:http://dx.doi.org/10.1016/j.jacc.2003.05.004
- ↑ Carlson, B. M. (1994). Human embryology and developmental biology. St. Louis: Mosby.
- ↑ Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999 Mar;126(6):1269-80. doi: 10.1242/dev.126.6.1269. PMID:10021345 doi:http://dx.doi.org/10.1242/dev.126.6.1269
- ↑ Furtado MB, Wilmanns JC, Chandran A, Tonta M, Biben C, Eichenlaub M, Coleman HA, Berger S, Bouveret R, Singh R, Harvey RP, Ramialison M, Pearson JT, Parkington HC, Rosenthal NA, Costa MW. A novel conditional mouse model for Nkx2-5 reveals transcriptional regulation of cardiac ion channels. Differentiation. 2016 Jan-Mar;91(1-3):29-41. doi: 10.1016/j.diff.2015.12.003., Epub 2016 Feb 17. PMID:26897459 doi:http://dx.doi.org/10.1016/j.diff.2015.12.003
- ↑ Durocher D, Chen CY, Ardati A, Schwartz RJ, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol. 1996 Sep;16(9):4648-55. doi: 10.1128/MCB.16.9.4648. PMID:8756621 doi:http://dx.doi.org/10.1128/MCB.16.9.4648
- ↑ Warren SA, Terada R, Briggs LE, Cole-Jeffrey CT, Chien WM, Seki T, Weinberg EO, Yang TP, Chin MT, Bungert J, Kasahara H. Differential role of Nkx2-5 in activation of the atrial natriuretic factor gene in the developing versus failing heart. Mol Cell Biol. 2011 Nov;31(22):4633-45. doi: 10.1128/MCB.05940-11. Epub 2011 Sep , 19. PMID:21930795 doi:http://dx.doi.org/10.1128/MCB.05940-11
- ↑ Pradhan L, Gopal S, Li S, Ashur S, Suryanarayanan S, Kasahara H, Nam HJ. Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5. Biochemistry. 2016 Mar 29;55(12):1702-10. doi: 10.1021/acs.biochem.6b00171. Epub , 2016 Mar 9. PMID:26926761 doi:http://dx.doi.org/10.1021/acs.biochem.6b00171
- ↑ Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997 Sep 15;16(18):5687-96. doi: 10.1093/emboj/16.18.5687. PMID:9312027 doi:http://dx.doi.org/10.1093/emboj/16.18.5687
- ↑ Wang J, Lu Y, Chen H, Yin M, Yu T, Fu Q. Investigation of somatic NKX2-5, GATA4 and HAND1 mutations in patients with tetralogy of Fallot. Pathology. 2011 Jun;43(4):322-6. doi: 10.1097/PAT.0b013e32834635a9. PMID:21519287 doi:http://dx.doi.org/10.1097/PAT.0b013e32834635a9
- ↑ Pradhan L, Gopal S, Li S, Ashur S, Suryanarayanan S, Kasahara H, Nam HJ. Intermolecular Interactions of Cardiac Transcription Factors NKX2.5 and TBX5. Biochemistry. 2016 Mar 29;55(12):1702-10. doi: 10.1021/acs.biochem.6b00171. Epub , 2016 Mar 9. PMID:26926761 doi:http://dx.doi.org/10.1021/acs.biochem.6b00171
- ↑ Lei X, Zhao J, Sagendorf JM, Rajashekar N, Xu J, Dantas Machado AC, Sen C, Rohs R, Feng P, Chen L. Crystal Structures of Ternary Complexes of MEF2 and NKX2-5 Bound to DNA Reveal a Disease Related Protein-Protein Interaction Interface. J Mol Biol. 2020 Sep 4;432(19):5499-5508. doi: 10.1016/j.jmb.2020.07.004. Epub, 2020 Jul 15. PMID:32681840 doi:http://dx.doi.org/10.1016/j.jmb.2020.07.004
- ↑ McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W597-600. doi:, 10.1093/nar/gkt376. Epub 2013 May 13. PMID:23671338 doi:http://dx.doi.org/10.1093/nar/gkt376
- ↑ Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing zebrafish heart. Anat Rec. 2000 Oct 1;260(2):148-57. doi:, 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. PMID:10993952 doi:<148::AID-AR50>3.0.CO;2-X http://dx.doi.org/10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X
- ↑ Harrington JK, Sorabella R, Tercek A, Isler JR, Targoff KL. Nkx2.5 is essential to establish normal heart rate variability in the zebrafish embryo. Am J Physiol Regul Integr Comp Physiol. 2017 Sep 1;313(3):R265-R271. doi:, 10.1152/ajpregu.00223.2016. Epub 2017 Jun 14. PMID:28615160 doi:http://dx.doi.org/10.1152/ajpregu.00223.2016
|



