Sandbox Reserved 1783
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
== Sodium Taurocholate Co-Transporting Peptide == | == Sodium Taurocholate Co-Transporting Peptide == | ||
<StructureSection load='7zyi' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='7zyi' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
== Background == | == Background == | ||
== Structural Overview == | == Structural Overview == | ||
| - | [[Image:Ntcp baby.png| | + | |
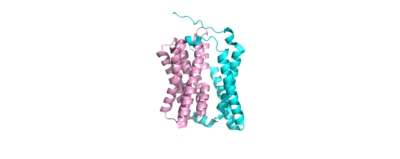
| + | There are two significant areas in the NTCP structure that facilitate ligand binding, which are referred to as "patches." Residues 84-87 of NTCP are patch 1, which are located on the TM2-TM3 loop in the core domain. This is also considered the extracellular region of NTCP “tunnel." Residues 157-165 NTCP are associated with patch 2. They are located on N-terminal half of the TM5 in the panel domain (residue sequence: KGIVISLVL). Patch 2 is also located in th extracellular region. These residues' importance was determined through mutations of these residues and examined through pull-down assays (Asami, et. al, 2022). | ||
| + | |||
| + | [[Image:Ntcp baby.png|400 px|right|thumb|Figure 1: NTCP separated by domains, a panel domain and a core domain involved in a conformational change]] | ||
<scene name='95/952711/Ntcp_regions_color_coded/1'>NTCP Separated by Domains</scene> | <scene name='95/952711/Ntcp_regions_color_coded/1'>NTCP Separated by Domains</scene> | ||
| + | |||
| + | |||
=== Binding Pocket === | === Binding Pocket === | ||
| + | |||
<scene name='95/952711/Sodium_residue_zoom_in_nctp/1'>Sodium binds to specific residues within the molecule. </scene> | <scene name='95/952711/Sodium_residue_zoom_in_nctp/1'>Sodium binds to specific residues within the molecule. </scene> | ||
<scene name='95/952711/Binding_site_2/1'>Binding Pocket 2</scene> | <scene name='95/952711/Binding_site_2/1'>Binding Pocket 2</scene> | ||
| + | |||
=== Conformation Change === | === Conformation Change === | ||
| + | |||
<scene name='95/952711/Open_pore_ntcp_non_transparent/1'>Open pore</scene> | <scene name='95/952711/Open_pore_ntcp_non_transparent/1'>Open pore</scene> | ||
<scene name='95/952711/Closed_pore_ntcp/1'>Closed Pore</scene> | <scene name='95/952711/Closed_pore_ntcp/1'>Closed Pore</scene> | ||
<scene name='95/952711/Open_pore_ntcp/1'>Open pore model</scene> | <scene name='95/952711/Open_pore_ntcp/1'>Open pore model</scene> | ||
<scene name='95/952711/Open_pore_with_bile_salts/1'>Open pore with Bile Salt</scene> | <scene name='95/952711/Open_pore_with_bile_salts/1'>Open pore with Bile Salt</scene> | ||
| + | |||
=== Mechanism === | === Mechanism === | ||
| + | |||
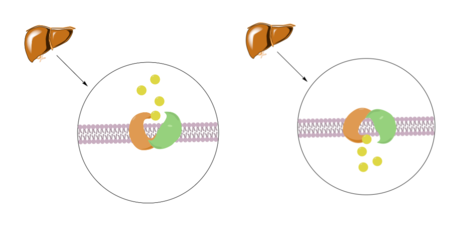
| + | [[Image:Screenshot_2023-03-20_at_3.59.09_PM.png|450 px|right|thumb|'''Figure 1.''' Bile Salt Uptake Mechanism.]] | ||
| + | |||
| + | The NTCP protein goes through a conformational change when assisting in the uptake of bile salt into the cell. This is accomplished through the opening of a wide transmembrane pore, creating a transport pathway for bile salts. The mechanism includes two sodium metal ions that allow for residue stabilization when going through the conformational change. Binding of the preS1 region of the HBV/HDV virus blocks any subsequent bile salt uptake. Thus, preS1 binding blocks the conformational change and entry of any salts into the cell. Residues 8-17 of preS1 are critical for NTCP:pres1 binding. Patch 1 and Patch 2 (external) residues interact with residues 8-17 of preS1 to facilitate binding. | ||
| + | |||
<scene name='95/952711/1_sodium_binding_to_ntcp/1'>One of the sodium molecules</scene> | <scene name='95/952711/1_sodium_binding_to_ntcp/1'>One of the sodium molecules</scene> | ||
| + | |||
== Significance == | == Significance == | ||
| - | + | NTCP serves a multitude of biological functions, including bile salt uptake and HBV/HDV binding. NTCP is a key player in the absorption and digestion of fats and fat-soluble vitamins in the body, as well as the synthesis of bile within the liver. The uptake of bile salts, transcriptional and post-transcriptional, are signaling molecules for the liver and intestine. | |
| + | |||
| + | === Bile Salt Uptake === | ||
| + | |||
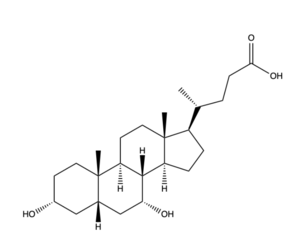
| + | [[Image:Bile Salt structure.png|300 px|left|thumb|'''Figure 2.''' Bile Salt Structure.]] | ||
| + | |||
| + | Disease of the liver is due to a decrease in bile salt uptake. This disease is transferred through bodily fluids between organisms. Liver Disease causes symptoms such as jaundice, abdominal pain and swelling, and swelling of the legs and feet. Liver disease can lead to liver cancer and other life-threatening diseases, making bile salt uptake essential to liver function. | ||
| - | + | === HBV/HDV === | |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 20:22, 20 March 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
Sodium Taurocholate Co-Transporting Peptide
| |||||||||||