This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1781
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
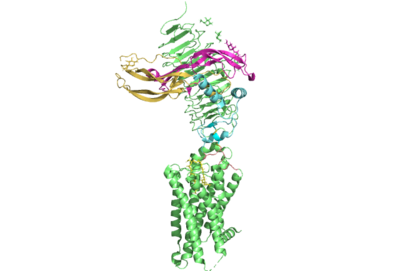
Several parts of TSHR are very important for the functioning of TSH signaling. The <scene name='95/952709/Hinge_region_real/2'>hinge region</scene> has been found to have impact on thyrotropin and autoantibody binding. However, the region is not required for the activation of the sensor. This is supported by the fact that select autoantibodies are able to bind to the activate the receptor without any contacts, change to the hinge region. This may be explained by the fact that | Several parts of TSHR are very important for the functioning of TSH signaling. The <scene name='95/952709/Hinge_region_real/2'>hinge region</scene> has been found to have impact on thyrotropin and autoantibody binding. However, the region is not required for the activation of the sensor. This is supported by the fact that select autoantibodies are able to bind to the activate the receptor without any contacts, change to the hinge region. This may be explained by the fact that | ||
== Relevance == | == Relevance == | ||
| - | + | <scene name='95/952709/Interactions_with_thyrotropin/1'>Glu 98 and Asp 91</scene> | |
== Structural highlights == | == Structural highlights == | ||
Revision as of 00:37, 29 March 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001 Jul;81(3):1097-142. doi: 10.1152/physrev.2001.81.3.1097. PMID: 11427693.
- ↑ Duan J, Xu P, Luan X, Ji Y, He X, Song N, Yuan Q, Jin Y, Cheng X, Jiang H, Zheng J, Zhang S, Jiang Y, Xu HE. Hormone- and antibody-mediated activation of the thyrotropin receptor. Nature. 2022 Aug 8. pii: 10.1038/s41586-022-05173-3. doi:, 10.1038/s41586-022-05173-3. PMID:35940204 doi:http://dx.doi.org/10.1038/s41586-022-05173-3
- ↑ Kohn LD, Shimura H, Shimura Y, Hidaka A, Giuliani C, Napolitano G, Ohmori M, Laglia G, Saji M. The thyrotropin receptor. Vitam Horm. 1995;50:287-384. doi: 10.1016/s0083-6729(08)60658-5. PMID: 7709602.
- ↑ . PMID:228484426