Introduction
Sodium-taurocholate Co-transporting Polypeptide (NTCP) is found within the membrane of hepatocytes, and its primary role is to facilitate the transport of bile salts into hepatocytes from the bloodstream. This is important because 90% of human bile salts are recycled daily, so the function of NTCP is critical in providing bile salts to solubilize fats for digestion. In addition to transporting bile salts into the cytoplasm of hepatocytes, NTCP also serves as a receptor for Hepatitis B (HBV) and Hepatitis D (HDV) viruses. (Insert 2D picture from the powerpoint for basic mechanism of both bile salt and HBV)
[1]
[2]
[3]
[4]
[5]
[6]
Structure
Structures were determined by cryogenic electron microscopy (Cryo-EM) of NTCP in complex with antibodies or nanobodies, revealing two key conformations in NTCP's transport mechanism. There are nine alpha helices spanning the membrane, with the N-terminus located on the extracellular side of the plasma membrane and the C-terminus located on the intracellular side. Transmembrane helices are connected by short loops as well as extracellular and intracellular alpha helices that lie nearly parallel to the membrane.
Domains
NTCP contains two characteristic domains: the core and panel domains. Movement of these two domains allows recognition and transport of bile salts into hepatocytes.
- Panel Domain: Residues 1-44, 155-208
- Formed by transmembrane helices TM1, TM5, and TM6.
- Core domain: Residues 45-154, 209-309
- Formed by the packing of a helix bundle of TM2, TM3, and TM4 with another helix bundle of TM7, TM8, and TM9. These two helix bundles are related by pseudo two-fold symmetry.
Proline/Glycine Hinge
Glycine and proline residues in the connecting loops and extra- and intracellular helices act as hinges in the mechanism of bile salt uptake. The flexibility allows separation of the core and panel domains, creating a pore open to the extracellular space and exposing critical Na+ binding sites. Once substrate binds the open-pore state, this hinge allows transition to close this pore relative to the extracellular side and open to the cytoplasmic side, thus allowing release of substrate into the cell.
Sodium Binding Sites
To transport a single bile salt from the blood to the cytoplasm of the hepatocyte, two sodium ions are required to be bound to to NTCP in the open-pore state. This is because the transport of bile salts into the cell is so thermodynamically unfavorable that the reaction has to be coupled to the favorable transport of two sodium into into the cell. It is thus an example of secondary active transport. When the bile salts are released into the cell, the protein is then found in the inward facing conformation, in which the pore through which the bile salt had just passed is now closed to the extracellular side. The residues interacting with the sodium ion in sodium binding site #1 include S105, N106, E257, and T123. The residues interacting with the sodium ion in sodium binding site #2 includes Q261 and Q68. Mutations to these significant residues inhibit the binding of sodium ions, and consequently, inhibit the transport of bile salts by NTCP.
- (self note - split up the two sodium binding sites so we have two green links instead of one)
Significant Residues
The majority of residues involved in bile salt uptake are also involved in HBV/HDV infection. (extracellular view) of Human NTCP have been shown to be vital for HBV/HDV virus recognition along with bile salt uptake. These residues were replaced in mice NTCP by human NTCP and conferred to successful binding of the virus. These residues are found in the extracellular loop connecting TM2 and TM3. (extracellular view) have also been shown to be vital for HBV/HDV viral recognition and bile salt uptake. These residues were mutated in monkey NTCP to the human residues and preS1 binding was then successful. These residues are found on the N-terminal end of TM5. The absence of residues in either of these hinders preS1 binding and therefore HBV/HDV infection. Interestingly, residues 84-87 do not affect bile acid uptake, so it is a potential site for blocking HBV/HDV infection while maintaining NTCP's ability to perform its normal function. Another important residue was discovered to be a single-nucleotide polymorphism in a small population in East Asia. , which is normally serine, being mutated to phenylalanine prevents preS1 binding and does not support bile acid transport. This residue is also found extracellularly, on TM8 of NTCP. There are 3 additional leucine residues that when mutated, block both preS1 binding and HBV/HDV infection. Replacing L27, L31, and L35 (INSERT GREEN LINK) with tryptophan residues presumably blocks the preS1 binding site preventing proper infection.
Function
Mechanism of Bile Salt Uptake
Bile salts recognize and bind to the . After binding, bile salts pass through the amphipathic pore (INSERT GREEN LINK). and NTCP transitions into the . In this conformation, the pore closes off relative to the extracellular side and opens to the cytoplasmic side. Transition to the inward facing state allows release of bile salts and sodium ions. It is not yet known how this transition exactly proceeds.
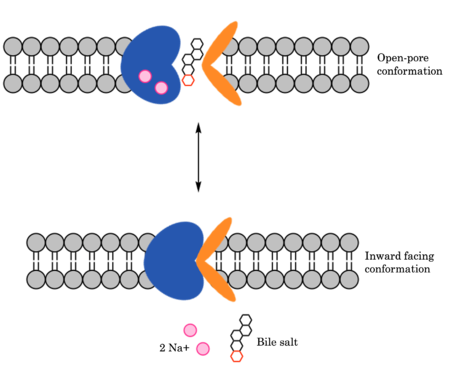
Mechanism of Bile Salt Uptake by NTCP.
Mechanism of HBV/HDV Infection
HBV and HDV viruses infect are transported through NTCP via secondary active transport. After binding to NTCP in the open-pore state, the viruses remain bound until low bile salt levels in the blood shift equilibria enough that endocytosis of NTCP occurs. Once in the cell, the viruses dissociate and infect. The exact mechanism of how HBV and HDV bind to NTCP is not certain, although two critical sites have been identified on NTCP: residues 84-87 and 157-165. Additionally, it has been shown that myristoylation of the HBV/HDV capsid is vital for recognition by NTCP, as well as residues 8-17 on HBV/HDV (sequence: NPLGFFPDHQ). (INSERT CITING) has proposed two mechanisms for how HBV/HDV binds to NTCP. The first proposes binding of the myristoyl group to the host cell membrane, while residues 8-17 interact with NTCP residues 157-165. The second proposes binding of the myristoyl group with residues 157-165 in the pore.
Medical Relevance
Bile salts are derived from cholesterol, and they serve an important role in the mechanical digestion of fats and ultimately facilitate the chemical digestion of lipids. The amphipathicity allows them to do this, solubilizing hydrophobic fats for transport in aqueous bodily fluids. Without bile salts, fats would spontaneously separate out of the aqueous solution in the duodenum and would not be accessible pancreatic lipase for breakdown. Proper fat digestion requires both pancreatic lipase and bile; thus, NTCP's function in recycling bile salts is critical.
Insight into NTCP's structure and function has implications for therapeutic treatment of HBV/HDV infection. For example, the inhibitory effect of Nb87 on myr-preS1 binding shows potential for therapeutics that stabilize NTCP inward-facing state as allosteric inhibitors of viral cell entry

