Sandbox Reserved 1794
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
=== Overview === | === Overview === | ||
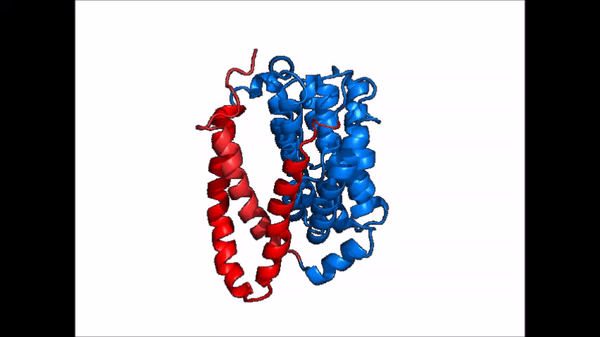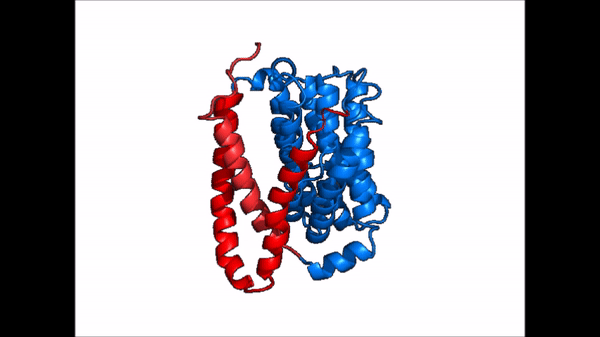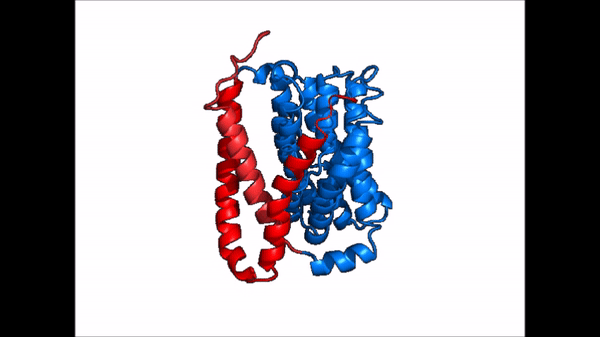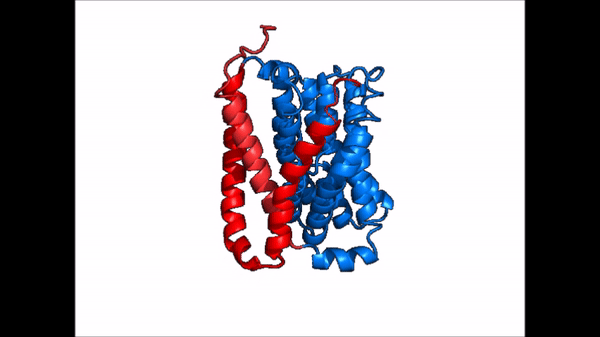
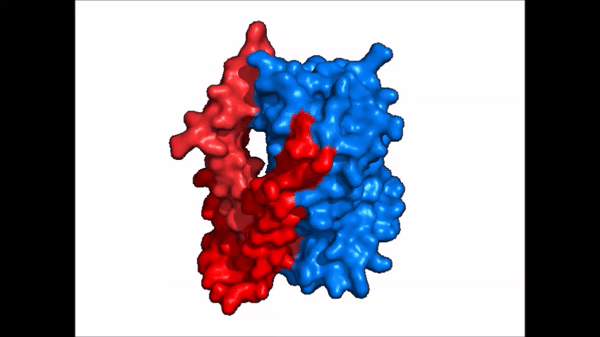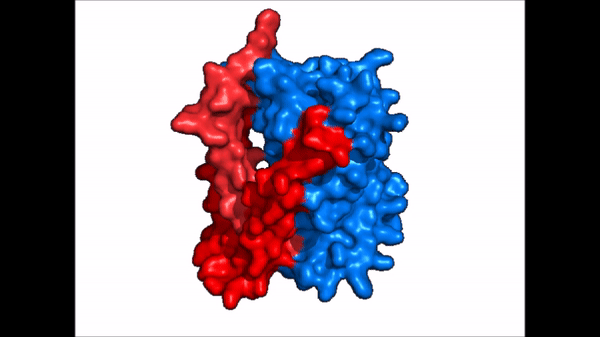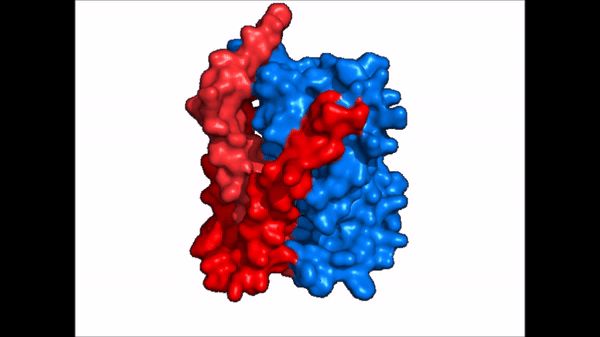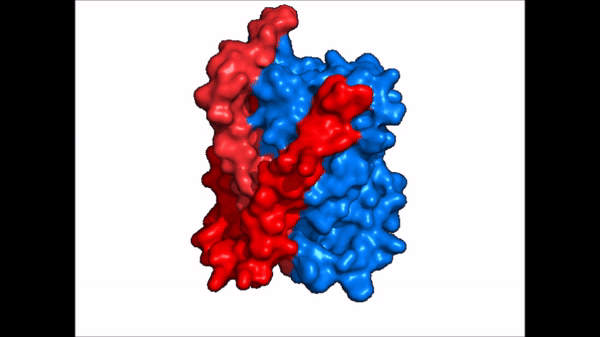
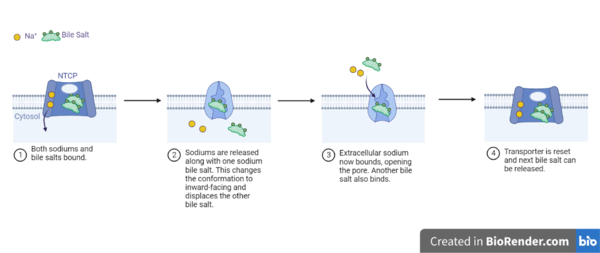
| - | NTCP is one continuous polypeptide chain consisting of a total of 9 transmembrane α helices (Fig. 2). The N-terminus of the polypeptide chain is found on the extracellular region of the plasma membrane while the C-terminus is located on the intracellular region. There are two distinct domains within the quaternary structure of NTCP: a <scene name='95/952722/Ntcp_core_domain-_blue/ | + | NTCP is one continuous polypeptide chain consisting of a total of 9 transmembrane α helices (Fig. 2). The N-terminus of the polypeptide chain is found on the extracellular region of the plasma membrane while the C-terminus is located on the intracellular region. There are two distinct domains within the quaternary structure of NTCP: a core domain and a , both being a part of the same polypeptide chain. The <scene name='95/952722/Ntcp_core_domain-_blue/6'>core domain</scene> <font color='#6060ff'><b>(blue)</b></font> includes 6 transmembrane α helices (TM2-4 and TM7-9) and demonstrates two-fold pseudosymmetry. The <scene name='95/952722/Ntcp_panel_domain-_red/2'>panel domain</scene> <font color='red'><b>(red)</b></font> consists of 3 transmembrane α helices (TM1 and TM5-6) and does not display symmetry. Within the core domain, there is a unique crossover between TM-3 and TM-8 that is known as the <scene name='95/952722/Ntcp_x_motif/5'>X motif</scene>. This motif is important because this is where the transporter's substrate binding site is located and within this motif lies essential residues that aid in the conformational change that NTCP undergoes. |
=== Binding Sites === | === Binding Sites === | ||
Revision as of 16:04, 5 April 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
Sodium Taurocholate Co-Transporting Polypeptide
| |||||||||||
References
- ↑ Stieger B. The role of the sodium-taurocholate cotransporting polypeptide (NTCP) and of the bile salt export pump (BSEP) in physiology and pathophysiology of bile formation. Handb Exp Pharmacol. 2011;(201):205-59. doi: 10.1007/978-3-642-14541-4_5. PMID: 21103971. DOI: DOI: 10.1007/978-3-642-14541-4_5.
- ↑ Geyer, J., Wilke, T. & Petzinger, E. The solute carrier family SLC10: more than a family of bile acid transporters regarding function and phylogenetic relationships. Naunyn Schmied Arch Pharmacol 372, 413–431 (2006). https://doi.org/10.1007/s00210-006-0043-8
- ↑ Park, JH., Iwamoto, M., Yun, JH. et al. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature 606, 1027–1031 (2022). https://doi.org/10.1038/s41586-022-04857-0.
- ↑ 4.0 4.1 Goutam, K., Ielasi, F.S., Pardon, E. et al. Structural basis of sodium-dependent bile salt uptake into the liver. Nature 606, 1015–1020 (2022). DOI: 10.1038/s41586-022-04723-z.
- ↑ Qi X. and Li W. (2022). Unlocking the secrets to human NTCP structure. The Innovation 3(5), 100294. https://doi.org/10.1016/j.xinn.2022.100294
Student Contributors
- Isabelle White
- Lena Barko