We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1790
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
=== SHOC2 === | === SHOC2 === | ||
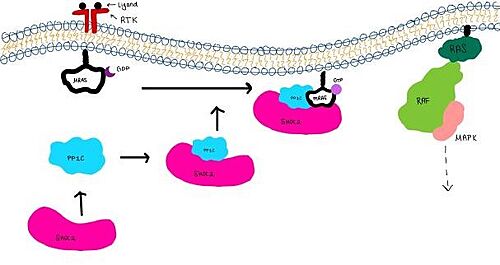
| - | <scene name='95/952717/Shoc2/1'>SHOC2</scene> | + | <scene name='95/952717/Shoc2/1'>SHOC2</scene> is a scaffold protein that is composed of 20 leucine-rich repeat domains that form a solenoid structure. The leucine rich region forms a concave hydrophobic core which is necessary for binding with PP1C and MRAS. |
=== PP1C === | === PP1C === | ||
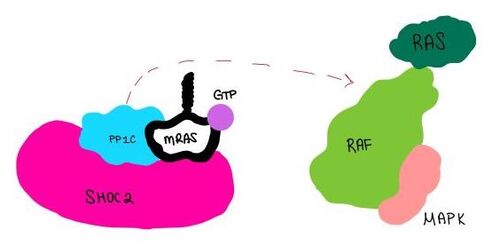
| - | <scene name='95/952717/Pp1c/1'>PP1C</scene> | + | <scene name='95/952717/Pp1c/1'>PP1C</scene> is a catalytic protein. After forming a ternary complex, the catalytic site on the protein interacts with Raf to acts as a phosphatase and dephosphorylates Ser 259. |
=== MRAS === | === MRAS === | ||
<scene name='95/952717/Mras/2'>MRAS</scene> is a membrane bound structure that aids the complex in localizing near other structures such as the RAS-RAF-MAPK complex in order to initiate downstream signaling. In its inactive state, MRAS is bound to GDP. When signaled by growth factors, the GDP is exchanged for GTP. The now <scene name='95/952718/Zoom_in_gtp/1'>GTP bound MRAS</scene> undergoes a conformational change of the switch I and switch II regions. This conformational change activates the protein allowing it to bind more easily with the SHOC2-PP1C complex. In comparison to other RAS proteins, MRAS has a greater affinity for the SHOC2-PP1C complex. | <scene name='95/952717/Mras/2'>MRAS</scene> is a membrane bound structure that aids the complex in localizing near other structures such as the RAS-RAF-MAPK complex in order to initiate downstream signaling. In its inactive state, MRAS is bound to GDP. When signaled by growth factors, the GDP is exchanged for GTP. The now <scene name='95/952718/Zoom_in_gtp/1'>GTP bound MRAS</scene> undergoes a conformational change of the switch I and switch II regions. This conformational change activates the protein allowing it to bind more easily with the SHOC2-PP1C complex. In comparison to other RAS proteins, MRAS has a greater affinity for the SHOC2-PP1C complex. | ||
| Line 30: | Line 30: | ||
=== SHOC2 and PP1C === | === SHOC2 and PP1C === | ||
| - | <scene name='95/952717/Shoc2_and_pp1c/1'>PP1C binds to SHOC2</scene> on its leucine rich region(LRR). Specifically, on two broad surfaces between LRR2 and LRR5 and between LRR7 and LRR11. Five main <scene name='95/952717/Shoc2_and_pp1c/2'>hydrogen bonds</scene> are made: E56-R182, E167-R203, E54-K180, R187-H178, R188-E155. The binding regions can also be shown as acidic and basic patches on <scene name='95/952718/Acid_base_pp1c/1'>PP1C</scene> and <scene name='95/952718/Acid_base_shoc2/1'>SHOC2</scene>. The corresponding patches interact to form a <scene name='95/952718/Acid_base_shoc2pp1c/1'>binary complex</scene>. | + | <scene name='95/952717/Shoc2_and_pp1c/1'>PP1C binds to SHOC2</scene> on its leucine rich region(LRR). Specifically, on two broad surfaces between LRR2 and LRR5 and between LRR7 and LRR11. Mutations made to the LRR were shown to completely inhibit the binding of PP1C. Five main <scene name='95/952717/Shoc2_and_pp1c/2'>hydrogen bonds</scene> are made: E56-R182, E167-R203, E54-K180, R187-H178, R188-E155. The binding regions can also be shown as acidic and basic patches on <scene name='95/952718/Acid_base_pp1c/1'>PP1C</scene> and <scene name='95/952718/Acid_base_shoc2/1'>SHOC2</scene>. The corresponding patches interact to form a <scene name='95/952718/Acid_base_shoc2pp1c/1'>binary complex</scene>. These interactions do not result in significant conformational changes. |
=== SHOC2 and MRAS === | === SHOC2 and MRAS === | ||
| - | MRAS is initially bound to GDP causing it to be in its inactive state. This form cannot bind to the SHOC2-PP1C complex due to steric clashing. Once GDP is exchanged for GTP to activate the protein, conformational | + | MRAS is initially bound to GDP causing it to be in its inactive state. This form cannot bind to the SHOC2-PP1C complex due to steric clashing. Once GDP is exchanged for GTP to activate the protein, conformational changes occur within the switch I and switch II regions to allow MRAS to interact with SHOC2. These interactions include hydrogen bonds and pi stacking. The primary hydrogen bonds are R288-Q71 and R177-E47. Pi staking occurs at R104-R83. |
=== PP1C and MRAS === | === PP1C and MRAS === | ||
| Line 39: | Line 39: | ||
== Signaling Pathway == | == Signaling Pathway == | ||
| - | [[Image:Signal_cascade_small.jpg| | + | [[Image:Signal_cascade_small.jpg|500 px|thumb|center|'''Figure 1:'''Signaling cascade is shown with SHOC2 in pink, PP1C in blue, and MRAs in white. ]] |
| - | [[Image:Dephosphorylation.jpg| | + | [[Image:Dephosphorylation.jpg|500 px|thumb|center|'''Figure 2:'''PP1C dephosphorylates RAF protein at serine 259 ]] |
Revision as of 19:26, 6 April 2023
This page, as it appeared on March 17, 2023, was featured in this article in the journal Biochemistry and Molecular Biology Education.
SHOC2-PP1C-MRAS
| |||||||||||