We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1793
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
==== Bile Salt ==== | ==== Bile Salt ==== | ||
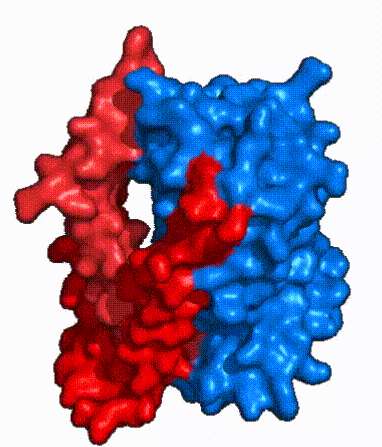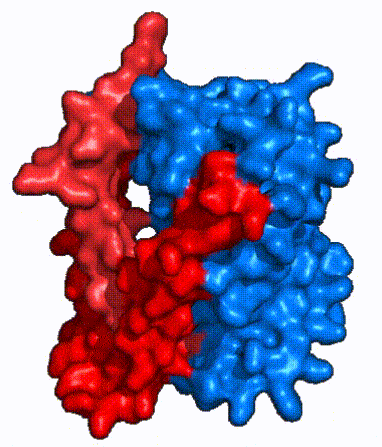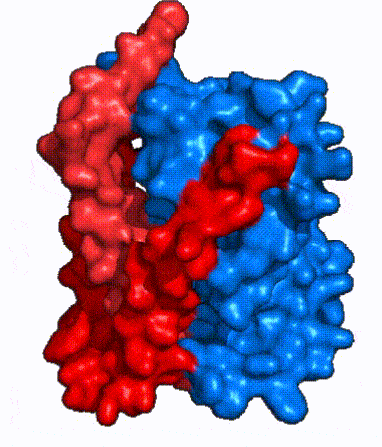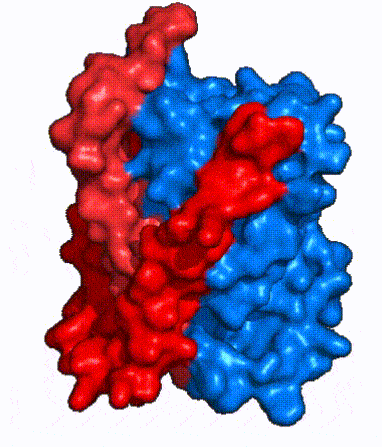
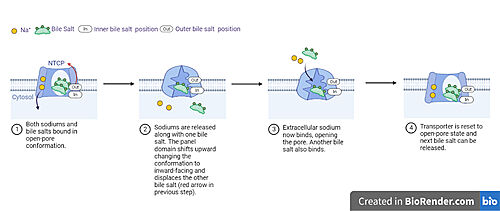
| - | The <scene name='95/952721/Amphipathic_patterns/2'>amphipathic pore</scene> is also characteristic of NTCP. The pore surface remains {{Template:ColorKey_Hydrophobic}}, while lining of the open pore state is largely {{Template:ColorKey_Polar}}. However, in the | + | The <scene name='95/952721/Amphipathic_patterns/2'>amphipathic pore</scene> is also characteristic of NTCP. The pore surface remains {{Template:ColorKey_Hydrophobic}}, while lining of the open pore state is largely {{Template:ColorKey_Polar}}. However, in the inward-facing, or closed-pore, conformation the polar pore residues are inaccessible. When the pore is closed only the surface hydrophobic residues are observed. As the pore opens up inner polar residues become accessible allowing for the binding of substrates. <Ref name = The pattern of hydrophobic and polar residues within the pore is believed to follow similar amphipathic patterns within taurocholate and other NTCP substrates, such as [https://en.wikipedia.org/wiki/Steroid steroids] and [https://en.wikipedia.org/wiki/Thyroid_hormones thyroid hormones]. <Ref name = Qi> Qi X. and Li W. (2022). Unlocking the secrets to human NTCP structure. The Innovation 3(5), 100294. https://doi.org/10.1016/j.xinn.2022.100294 </ref> Thus the channel provides specificity while preventing leakage of other substrates. When observing the relevant <scene name='95/952722/Bile_salts_res/1'>bile salt binding residues</scene> it is shown that some residues form Van der Waals interactions while others will form dipole-dipole or ionic interactions with bile salt substrates. The core domain appears to contribute most of the polar domains, while the panel domain contributes more hydrophobic residues. |
== Conformational Change == | == Conformational Change == | ||
{| | {| | ||
Revision as of 18:08, 7 April 2023
Sodium Bile Salt Co-Transporting Protein
| |||||||||||
References
- ↑ Stieger B. The role of the sodium-taurocholate cotransporting polypeptide (NTCP) and of the bile salt export pump (BSEP) in physiology and pathophysiology of bile formation. Handb Exp Pharmacol. 2011;(201):205-59. doi: 10.1007/978-3-642-14541-4_5. PMID: 21103971. DOI: DOI: 10.1007/978-3-642-14541-4_5.
- ↑ Geyer, J., Wilke, T. & Petzinger, E. The solute carrier family SLC10: more than a family of bile acid transporters regarding function and phylogenetic relationships. Naunyn Schmied Arch Pharmacol 372, 413–431 (2006). https://doi.org/10.1007/s00210-006-0043-8
- ↑ 3.0 3.1 3.2 Park, JH., Iwamoto, M., Yun, JH. et al. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature 606, 1027–1031 (2022). https://doi.org/10.1038/s41586-022-04857-0.
- ↑ 4.0 4.1 Goutam, K., Ielasi, F.S., Pardon, E. et al. Structural basis of sodium-dependent bile salt uptake into the liver. Nature 606, 1015–1020 (2022). DOI: 10.1038/s41586-022-04723-z.
- ↑ Qi X. and Li W. (2022). Unlocking the secrets to human NTCP structure. The Innovation 3(5), 100294. https://doi.org/10.1016/j.xinn.2022.100294
- ↑ 6.0 6.1 6.2 Liu, H., Irobalieva, R.N., Bang-Sørensen, R. et al. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res 32, 773–776 (2022). https://doi.org/10.1038/s41422-022-00680-4
- ↑ Latorraca, N. R.; Fastman, N. M.; Venkatakrishnan, A. J.; Frommer, W. B.; Dror, R. O.; Feng, L. Mechanism of Substrate Translocation in an Alternating Access Transporter. Cell 2017, 169 (1), 96–107.
- ↑ 8.0 8.1 8.2 8.3 Asami, J., Kimura, K.T., Fujita-Fujiharu, Y. et al. Structure of the bile acid transporter and HBV receptor NTCP. Nature 606, 1021–1026 (2022). https://doi.org/10.1038/s41586-022-04845-4
- ↑ Grove, J.; Marsh, M. The Cell Biology of Receptor-Mediated Virus Entry. Journal of Cell Biology 2011, 195 (7), 1071–1082.
- ↑ 10.0 10.1 Herrscher C, Roingeard P, Blanchard E. Hepatitis B Virus Entry into Cells. Cells. 2020 Jun 18;9(6):1486. doi: 10.3390/cells9061486. PMID: 32570893; PMCID: PMC7349259.