We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1771
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
===Antigen Binding Site=== | ===Antigen Binding Site=== | ||
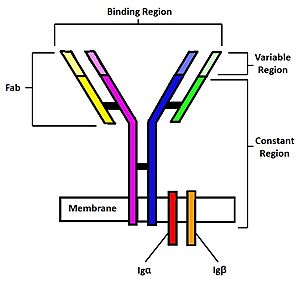
| - | [[Image: | + | [[Image:BCR Diagram2.jpg | 300 px|thumb | right| '''Figure 2.''' Diagram of the human B Cell Receptor with the Fab fragments, binding region, variable regions, constant regions and Iga/ Igb labelled. Heavy chain 1 is represented in blue, heavy chain 2 in magenta, light chain 1 in green, and light chain 2 in yellow. Disulfide bridges are represented with solid black lines connecting the heavy chains to themselves and to the light chains.]] |
The binding of an antigen to the human B Cell receptor is identical to other common soluble antibodies (such as [https://en.wikipedia.org/wiki/Immunoglobulin_G IgG], [https://en.wikipedia.org/wiki/Immunoglobulin_A IgA], [https://en.wikipedia.org/wiki/Immunoglobulin_M IgM], [https://en.wikipedia.org/wiki/Immunoglobulin_E IgE], or [https://en.wikipedia.org/wiki/Immunoglobulin_D IgD]). The antibody portion of the B Cell Receptor is roughly "Y" shaped and consists of two identical <scene name='95/952701/Heavy_chains_highlight/2'>heavy</scene> and two identical <scene name='95/952701/Light_chains_highlight/3'>light</scene> chains creating two similar epitope binding regions<Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref> (figure 1). Two antigen molecules can bind independent of one another to produce a response. Matching with standard FABs, the Ig portion has constant and variable region. The stem of the "Y" is a <scene name='95/952701/Constant_stem/1'>constant region</scene> ([https://en.wikipedia.org/wiki/Antibody#CDRs,_Fv,_Fab_and_Fc_Regions Fc]) composed of only heavy chain interactions<Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref>. The two heavy chains then branch at a flexible <scene name='95/952701/Hinge/1'>hinge region</scene>. These interact individually with one light chain creating two [https://en.wikipedia.org/wiki/Antibody#CDRs,_Fv,_Fab_and_Fc_Regions Fab] fragments or branches of the "Y"<Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref>. Light and heavy chains are held together via weak intermolecular forces and disulfide bridges<Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref>. Each <scene name='95/952701/Fab/1'>Fab fragment</scene> then terminates with two <scene name='95/952701/Fv_region/1'>variable regions</scene> ([https://en.wikipedia.org/wiki/Antibody#CDRs,_Fv,_Fab_and_Fc_Regions Fv]). These variable regions consist of hyper-variable loops, desired random coils of amino acids selected for specific recognition of a desired antigen <Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref>. Binding to an antigen is determined based on intermolecular interactions to the hyper variable loops, and selectivity is provided by unique hyper variable loop sequences. Due to the identical structure of Fab fragments, BCR will recognize antigens in the same manner as do free antibodies. This is emphasized by Ma ''et al.'' who studied the IgG- BCR (VRC01) that targets gp120 of the HIV-1 virus. By comparing structures of membrane bound and free antibodies, the membrane bound BCR form has an identical structure to the free antibody suggesting that they recognize antigens in the same way. <Ref name="Ma X">Ma X, Zhu Y, Dong D, Chen Y, Wang S, Yang D, Ma Z, Zhang A, Zhang F, Guo C, Huang Z. Cryo-EM structures of two human B cell receptor isotypes. Science. 2022 Aug 19;377(6608):880-885. [doi: 10.1126/science.abo3828. Epub 2022 Aug 18. PMID: 35981028]</Ref> This leads to a conformational change in the protein and transmits the signal through the membrane. <Ref name="Shen Z."> Zhixun Shen, Sichen Liu, Xinxin Li, Zhengpeng Wan, Youxiang Mao, Chunlai Chen, Wanli Liu (2019) Conformational change within the extracellular domain of B cell receptor in B cell activation upon antigen binding eLife 8:e42271. Doi: https://doi.org/10.7554/eLife.42271 </Ref> | The binding of an antigen to the human B Cell receptor is identical to other common soluble antibodies (such as [https://en.wikipedia.org/wiki/Immunoglobulin_G IgG], [https://en.wikipedia.org/wiki/Immunoglobulin_A IgA], [https://en.wikipedia.org/wiki/Immunoglobulin_M IgM], [https://en.wikipedia.org/wiki/Immunoglobulin_E IgE], or [https://en.wikipedia.org/wiki/Immunoglobulin_D IgD]). The antibody portion of the B Cell Receptor is roughly "Y" shaped and consists of two identical <scene name='95/952701/Heavy_chains_highlight/2'>heavy</scene> and two identical <scene name='95/952701/Light_chains_highlight/3'>light</scene> chains creating two similar epitope binding regions<Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref> (figure 1). Two antigen molecules can bind independent of one another to produce a response. Matching with standard FABs, the Ig portion has constant and variable region. The stem of the "Y" is a <scene name='95/952701/Constant_stem/1'>constant region</scene> ([https://en.wikipedia.org/wiki/Antibody#CDRs,_Fv,_Fab_and_Fc_Regions Fc]) composed of only heavy chain interactions<Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref>. The two heavy chains then branch at a flexible <scene name='95/952701/Hinge/1'>hinge region</scene>. These interact individually with one light chain creating two [https://en.wikipedia.org/wiki/Antibody#CDRs,_Fv,_Fab_and_Fc_Regions Fab] fragments or branches of the "Y"<Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref>. Light and heavy chains are held together via weak intermolecular forces and disulfide bridges<Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref>. Each <scene name='95/952701/Fab/1'>Fab fragment</scene> then terminates with two <scene name='95/952701/Fv_region/1'>variable regions</scene> ([https://en.wikipedia.org/wiki/Antibody#CDRs,_Fv,_Fab_and_Fc_Regions Fv]). These variable regions consist of hyper-variable loops, desired random coils of amino acids selected for specific recognition of a desired antigen <Ref name="Janeway CA">Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. </Ref>. Binding to an antigen is determined based on intermolecular interactions to the hyper variable loops, and selectivity is provided by unique hyper variable loop sequences. Due to the identical structure of Fab fragments, BCR will recognize antigens in the same manner as do free antibodies. This is emphasized by Ma ''et al.'' who studied the IgG- BCR (VRC01) that targets gp120 of the HIV-1 virus. By comparing structures of membrane bound and free antibodies, the membrane bound BCR form has an identical structure to the free antibody suggesting that they recognize antigens in the same way. <Ref name="Ma X">Ma X, Zhu Y, Dong D, Chen Y, Wang S, Yang D, Ma Z, Zhang A, Zhang F, Guo C, Huang Z. Cryo-EM structures of two human B cell receptor isotypes. Science. 2022 Aug 19;377(6608):880-885. [doi: 10.1126/science.abo3828. Epub 2022 Aug 18. PMID: 35981028]</Ref> This leads to a conformational change in the protein and transmits the signal through the membrane. <Ref name="Shen Z."> Zhixun Shen, Sichen Liu, Xinxin Li, Zhengpeng Wan, Youxiang Mao, Chunlai Chen, Wanli Liu (2019) Conformational change within the extracellular domain of B cell receptor in B cell activation upon antigen binding eLife 8:e42271. Doi: https://doi.org/10.7554/eLife.42271 </Ref> | ||
Revision as of 04:55, 14 April 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
IgM B-cell Receptor
| |||||||||||
References
- ↑ Robinson R. Distinct B cell receptor functions are determined by phosphorylation. PLoS Biol. 2006 Jul;4(7):e231. doi: 10.1371/journal.pbio.0040231. Epub 2006 May 30. PMID: 20076604; PMCID: PMC1470464.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Su Q, Chen M, Shi Y, Zhang X, Huang G, Huang B, Liu D, Liu Z, Shi Y. Cryo-EM structure of the human IgM B cell receptor. Science. 2022 Aug 19;377(6608):875-880. [doi: 10.1126/science.abo3923. Epub 2022 Aug 18. PMID: 35981043.]
- ↑ 3.0 3.1 3.2 3.3 3.4 Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001.
- ↑ Ma X, Zhu Y, Dong D, Chen Y, Wang S, Yang D, Ma Z, Zhang A, Zhang F, Guo C, Huang Z. Cryo-EM structures of two human B cell receptor isotypes. Science. 2022 Aug 19;377(6608):880-885. [doi: 10.1126/science.abo3828. Epub 2022 Aug 18. PMID: 35981028]
- ↑ Zhixun Shen, Sichen Liu, Xinxin Li, Zhengpeng Wan, Youxiang Mao, Chunlai Chen, Wanli Liu (2019) Conformational change within the extracellular domain of B cell receptor in B cell activation upon antigen binding eLife 8:e42271. Doi: https://doi.org/10.7554/eLife.42271
- ↑ 6.0 6.1 Tolar P, Pierce SK. Unveiling the B cell receptor structure. Science. 2022 Aug 19;377(6608):819-820. [doi: 10.1126/science.add8065. Epub 2022 Aug 18. PMID: 35981020.]
- ↑ Althwaiqeb, S. Histology, B Cell Lymphocyte; StatPearls Publishing, 2023.
- ↑ 8.0 8.1 8.2 Yanaba K, Bouaziz JD, Matsushita T, Magro CM, St Clair EW, Tedder TF. B-lymphocyte contributions to human autoimmune disease. Immunol Rev. 2008 Jun;223:284-99. doi: 10.1111/j.1600-065X.2008.00646.x. PMID: 18613843.
- ↑ 9.0 9.1 Chandrashekara S. The treatment strategies of autoimmune disease may need a different approach from conventional protocol: a review. Indian J Pharmacol. 2012 Nov-Dec;44(6):665-71. doi: 10.4103/0253-7613.103235. PMID: 23248391; PMCID: PMC3523489.
- ↑ Shu SA, Wang J, Tao MH, Leung PS. Gene Therapy for Autoimmune Disease. Clin Rev Allergy Immunol. 2015 Oct;49(2):163-76. doi: 10.1007/s12016-014-8451-x. PMID: 25277817.
Student Contributors
- Joel Wadas
- Olivia Gooch
- Delaney Lupoi