We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1794
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
=== Overview === | === Overview === | ||
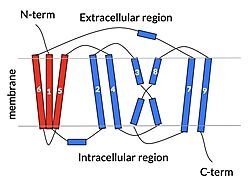
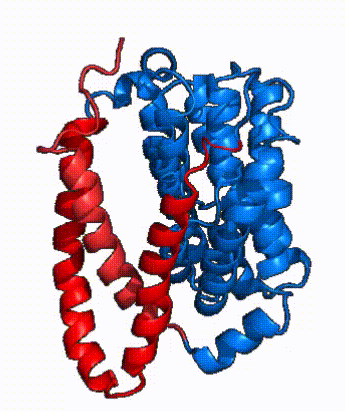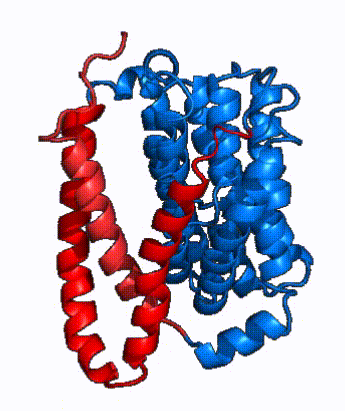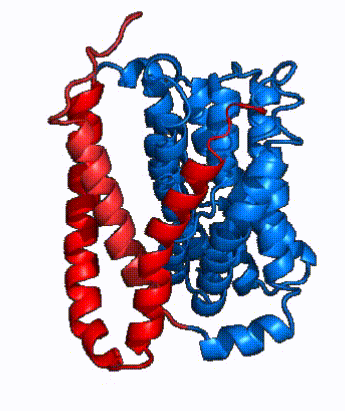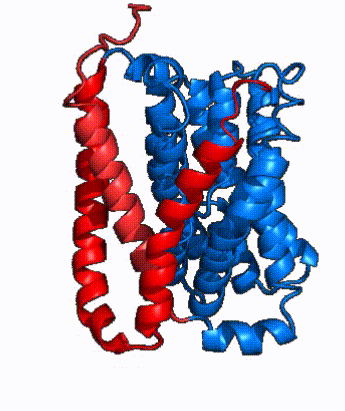
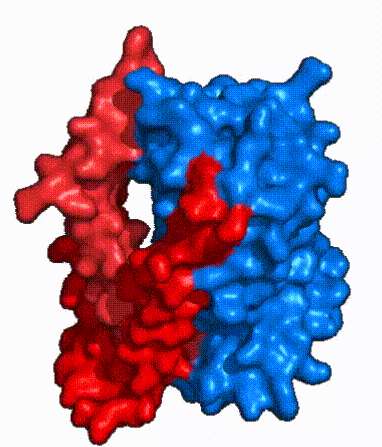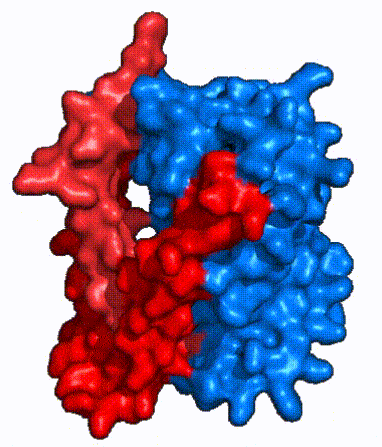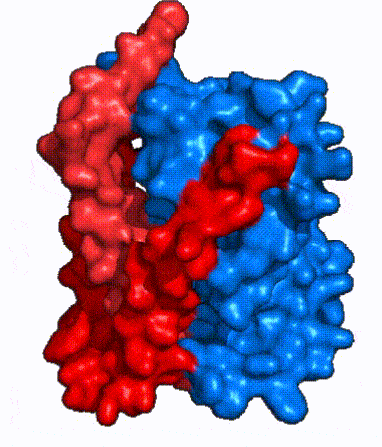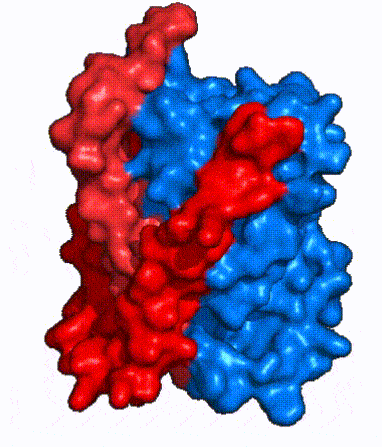
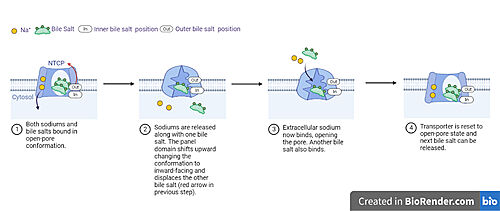
| - | NTCP is one continuous polypeptide chain containing <scene name='95/952722/Labeled_9_helices/5'>9 transmembrane alpha helices</scene>.<ref name="Goutam"/> The N-terminus of the polypeptide chain extrudes into the extracellular region of the plasma membrane while the C-terminus juts into the intracellular region. NTCP contains <scene name='95/952722/Ntcp_core_domain-_blue/10'>Two distinct sub domains</scene>: a core domain and a panel domain, which together channel opening and bile salt transport (Fig. 2). The <scene name='95/952722/Ntcp_core_domain-_blue/8'>core domain</scene> <font color='#6060ff'><b>(blue)</b></font> contains 6 transmembrane α helices (TM2-4 and TM7-9) and demonstrates [https://en.wikipedia.org/wiki/Protein_structure two-fold pseudosymmetry]. The <scene name='95/952722/Ntcp_panel_domain-_red/4'>panel domain</scene> <font color='red'><b>(red)</b></font> consists of 3 transmembrane α helices (TM1 and TM5-6) and is asymmetrical. Within the core domain, a unique crossover between TM-3 and TM-8 creates an <scene name='95/952722/Ntcp_x_motif/14'>X motif</scene>. The X motif contains the substrate binding site and essential residues for the conformational change required for transport. The core and panel domains are also connected by both extracellular and intracellular <scene name='95/952722/Connector_helices/6'>connector helices</scene> that are separate from the | + | NTCP is one continuous polypeptide chain containing <scene name='95/952722/Labeled_9_helices/5'>9 transmembrane alpha helices</scene>.<ref name="Goutam"/> The N-terminus of the polypeptide chain extrudes into the extracellular region of the plasma membrane while the C-terminus juts into the intracellular region. NTCP contains <scene name='95/952722/Ntcp_core_domain-_blue/10'>Two distinct sub domains</scene>: a core domain and a panel domain, which together channel opening and bile salt transport (Fig. 2). The <scene name='95/952722/Ntcp_core_domain-_blue/8'>core domain</scene> <font color='#6060ff'><b>(blue)</b></font> contains 6 transmembrane α helices (TM2-4 and TM7-9) and demonstrates [https://en.wikipedia.org/wiki/Protein_structure two-fold pseudosymmetry]. The <scene name='95/952722/Ntcp_panel_domain-_red/4'>panel domain</scene> <font color='red'><b>(red)</b></font> consists of 3 transmembrane α helices (TM1 and TM5-6) and is asymmetrical. Within the core domain, a unique crossover between TM-3 and TM-8 creates an <scene name='95/952722/Ntcp_x_motif/14'>X motif</scene>. The X motif contains the substrate binding site and essential residues for the conformational change required for transport. The core and panel domains are also connected by both extracellular and intracellular <scene name='95/952722/Connector_helices/6'>connector helices</scene> that are separate from the <scene name='95/952722/Labeled_9_helices/5'>9 transmembrane alpha helices</scene>. |
Revision as of 02:47, 17 April 2023
Sodium Taurocholate Co-Transporting Polypeptide
| |||||||||||
References
- ↑ Stieger B. The role of the sodium-taurocholate cotransporting polypeptide (NTCP) and of the bile salt export pump (BSEP) in physiology and pathophysiology of bile formation. Handb Exp Pharmacol. 2011;(201):205-59. doi: 10.1007/978-3-642-14541-4_5. PMID: 21103971. DOI: DOI: 10.1007/978-3-642-14541-4_5.
- ↑ Geyer, J., Wilke, T. & Petzinger, E. The solute carrier family SLC10: more than a family of bile acid transporters regarding function and phylogenetic relationships. Naunyn Schmied Arch Pharmacol 372, 413–431 (2006). https://doi.org/10.1007/s00210-006-0043-8
- ↑ 3.0 3.1 3.2 Park, JH., Iwamoto, M., Yun, JH. et al. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature 606, 1027–1031 (2022). https://doi.org/10.1038/s41586-022-04857-0.
- ↑ 4.0 4.1 4.2 4.3 4.4 Goutam, K., Ielasi, F.S., Pardon, E. et al. Structural basis of sodium-dependent bile salt uptake into the liver. Nature 606, 1015–1020 (2022). DOI: 10.1038/s41586-022-04723-z.
- ↑ Qi X. and Li W. (2022). Unlocking the secrets to human NTCP structure. The Innovation 3(5), 100294. https://doi.org/10.1016/j.xinn.2022.100294
- ↑ 6.0 6.1 6.2 Liu, H., Irobalieva, R.N., Bang-Sørensen, R. et al. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res 32, 773–776 (2022). https://doi.org/10.1038/s41422-022-00680-4
- ↑ Latorraca, N. R.; Fastman, N. M.; Venkatakrishnan, A. J.; Frommer, W. B.; Dror, R. O.; Feng, L. Mechanism of Substrate Translocation in an Alternating Access Transporter. Cell 2017, 169 (1), 96–107.
- ↑ 8.0 8.1 8.2 8.3 Asami, J., Kimura, K.T., Fujita-Fujiharu, Y. et al. Structure of the bile acid transporter and HBV receptor NTCP. Nature 606, 1021–1026 (2022). https://doi.org/10.1038/s41586-022-04845-4
- ↑ Grove, J.; Marsh, M. The Cell Biology of Receptor-Mediated Virus Entry. Journal of Cell Biology 2011, 195 (7), 1071–1082.
- ↑ 10.0 10.1 Herrscher C, Roingeard P, Blanchard E. Hepatitis B Virus Entry into Cells. Cells. 2020 Jun 18;9(6):1486. doi: 10.3390/cells9061486. PMID: 32570893; PMCID: PMC7349259.
Student Contributors
- Isabelle White
- Lena Barko