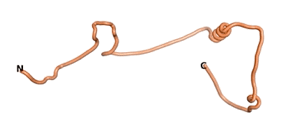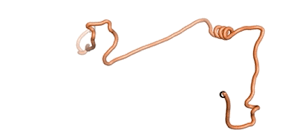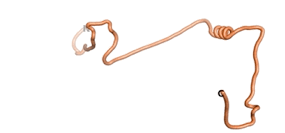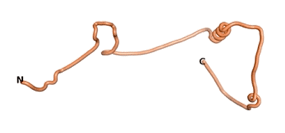| Do ‘Newly Born’ Orphan Proteins Resemble ‘Never Born’ Proteins? A Study Using Three Deep Learning Algorithms
Jing Liu, Rongqing Yuan, Wei Shao, Jitong Wang, Israel Silman and Joel Sussman [1]
Newly Born proteins, or orphan proteins, have no sequence homology to other proteins and occur in single species or within a taxonomically restricted gene (TRG) family.
Never Born proteins are random polypeptides with amino acid content similar to that of native proteins.
Can recently developed AI/Deep Learning tools for predicting 3D protein structures be useful to see if Newly Born proteins are similar to Never Born proteins? Three of the most widely used tools are:
- AlphaFold2 (AF2) [2]
- RoseTTAFold (RTF) [3]
- Evolutionary Scale Modeling (ESM-2)[4]
AF2 and RTF predict, by default, five top models, while ESM-2 predicts only one model. Morphing between the top models of AF2 and those of RTF gives a visual feeling of how similar these 5 models are for each method.
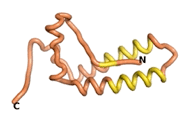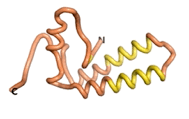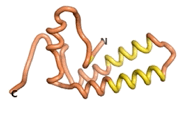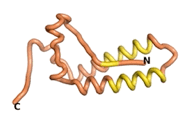
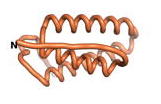
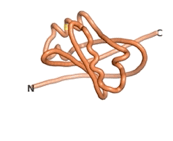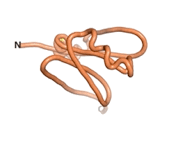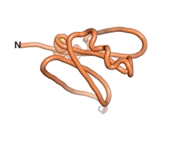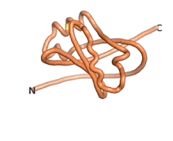
True orphan proteins have no sequence homology to any existing protein. We thought, therefore, that the Never Born proteins generated and investigated by Tretyachenko et al.[5] would serve as a valuable benchmark for comparison. In their study, they experimentally showed that some Never Born proteins folded into compact structures, e.g., as seen for Sequences #1856 and #6387.
| RTF-1856
| ESM-1856
| AF2-1856
|
| 
| 
|
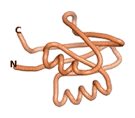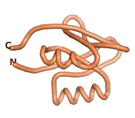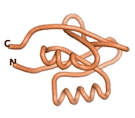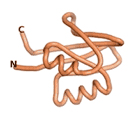
| RTF-6387
| ESM-6387
| AF2-6387
|
| 
| 
|
Other never born proteins experimentally appear to belong to the category of intrinsically disordered proteins (IDPs)[6], e.g., as seen for Sequence #3703.
| RTF-3703
| ESM-3703
| AF2-3703
|
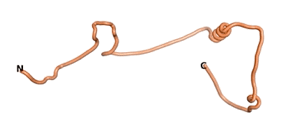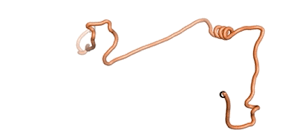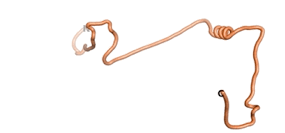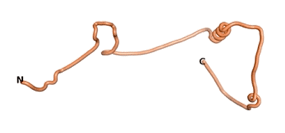
| 
| 
|
We then went on to use the three algorithms on orphan proteins and taxonomically restricted gene products (TRGP) for which no experimental structures were available. We did this in order to see how the predictions of the three algorithms would compare, and whether they would predict novel folds. Although many ORFs have been identified that code for putative orphan proteins, only in a limited number of cases has their association with a well-defined biological activity been established. We have identified seven such proteins for which the necessary sequence data are also available. The number of amino acids for these seven orphans/TRGPs ranges from 109 to 632.
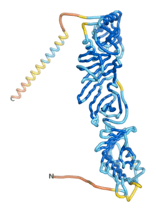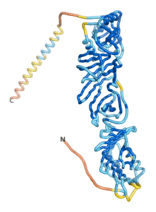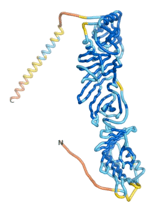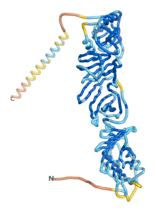
As an initial step in characterizing these seven proteins, we utilized FoldIndex[7] and flDPnn[8] to investigate whether they were predicted to be intrinsically disordered proteins (IDP) or folded. Five of the proteins are predicted to be almost completely folded, while the other two, TaFROG and Newtic1, are classified as IDPs since they are predicted to be disordered throughout almost their entire sequences. Of the seven proteins studied, only HCO_011565, a 632 residue nematode protein that was shown to be the target of the nematodicidal small molecule, appears to be fully folded. The three algorithms predict almost identical structures as well as very high pLDDT scores. Most likely, this is for two reasons. Firstly, rather than being a true orphan, HCO_011565 is the product of a TRG[9], with the BLAST search has revealed that the first 74 homologous sequences, with the lowest E values, were all from nematodes. Secondly, the DALI server revealed a number of hits for the entire predicted structure, as well as for the three subdomains predicted by all three algorithms.
| RTF-HCO_011565
| ESM-HCO_011565
| AF2-HCO_011565
|
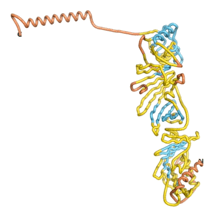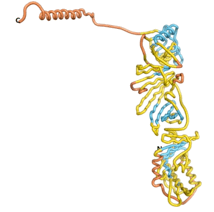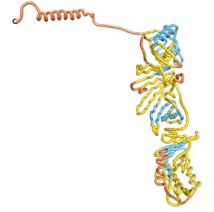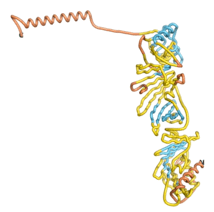
| 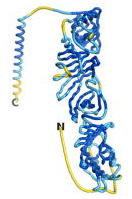
| 
|
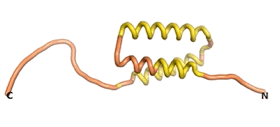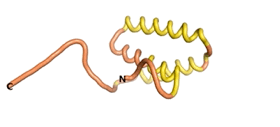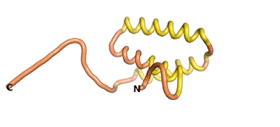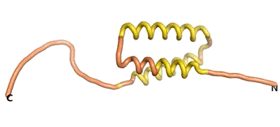
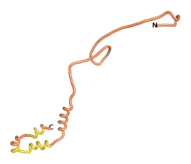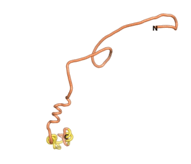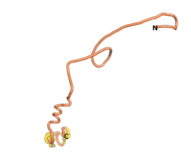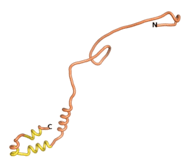
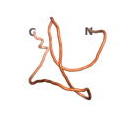
An example of an Orphan protein that is predicted by all three algorithms to be an IDP is the wheat protein TaFROG. It contains 130 amino acids localized in the nucleus. It confers resistance on wheat to the mycotoxigenic fungus, Fusarium graminearum[10].
| RTF-TaFROG
| ESM-TaFROG
| AF2-TaFROG
|
| 
| 
|
Although the pLDDT[11] scores for most of the Never Born and Newly Born proteins are low, the overall conformation of the 3D structures that they predict appear to give very consistent correlations of a compact folded protein versus an IDP. Thus, the approach to using recently developed AI/Deep Learning tools to indicate the overall shape of these proteins is useful.
Because the sequences of orphan proteins lack homology information, protein structure prediction for them has recently become a hot topic[12]. The approaches and methodologies we have implemented in this study may provide a starting point for datasets and protocols to evaluate the performance of structure prediction algorithms on sequences that lack homology to other sequences. It has not escaped our notice that this topic may significantly impact our understanding of how new traits evolve from orphan proteins.
References
- ↑ Liu J, Yuan R, Shao W, Wang J, Silman I, Sussman JL. Do "Newly Born" orphan proteins resemble "Never Born" proteins? A study using three deep learning algorithms. Proteins. 2023 Apr 24. PMID:37092778 doi:10.1002/prot.26496
- ↑ Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Jul 15. pii: 10.1038/s41586-021-03819-2. doi:, 10.1038/s41586-021-03819-2. PMID:34265844 doi:http://dx.doi.org/10.1038/s41586-021-03819-2
- ↑ Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millan C, Park H, Adams C, Glassman CR, DeGiovanni A, Pereira JH, Rodrigues AV, van Dijk AA, Ebrecht AC, Opperman DJ, Sagmeister T, Buhlheller C, Pavkov-Keller T, Rathinaswamy MK, Dalwadi U, Yip CK, Burke JE, Garcia KC, Grishin NV, Adams PD, Read RJ, Baker D. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021 Jul 15. pii: science.abj8754. doi: 10.1126/science.abj8754. PMID:34282049 doi:http://dx.doi.org/10.1126/science.abj8754
- ↑ Lin Z et al. & Rives, A (2022) Language models of protein sequences at the scale of evolution enable accurate structure prediction. bioRxiv. 2022:2022.2007.2020.500902. DOI:10.1101/2022.07.20.500902
- ↑ Tretyachenko V, Vymětal J, Bednárová L, Kopecký V Jr, Hofbauerová K, Jindrová H, Hubálek M, Souček R, Konvalinka J, Vondrášek J, Hlouchová K. Random protein sequences can form defined secondary structures and are well-tolerated in vivo. Sci Rep. 2017 Nov 13;7(1):15449. PMID:29133927 doi:10.1038/s41598-017-15635-8
- ↑ Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008 Dec;18(6):756-64. Epub 2008 Nov 17. PMID:18952168 doi:10.1016/j.sbi.2008.10.002
- ↑ Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005 Aug 15;21(16):3435-8. Epub 2005 Jun 14. PMID:15955783 doi:http://dx.doi.org/10.1093/bioinformatics/bti537
- ↑ Hu G, Katuwawala A, Wang K, Wu Z, Ghadermarzi S, Gao J, Kurgan L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat Commun. 2021 Jul 21;12(1):4438. PMID:34290238 doi:10.1038/s41467-021-24773-7
- ↑ Taki AC, Wang T, Nguyen NN, Ang CS, Leeming MG, Nie S, Byrne JJ, Young ND, Zheng Y, Ma G, Korhonen PK, Koehler AV, Williamson NA, Hofmann A, Chang BCH, Häberli C, Keiser J, Jabbar A, Sleebs BE, Gasser RB. Thermal proteome profiling reveals Haemonchus orphan protein HCO_011565 as a target of the nematocidal small molecule UMW-868. Front Pharmacol. 2022 Oct 14;13:1014804. PMID:36313370 doi:10.3389/fphar.2022.1014804
- ↑ Perochon A, Jianguang J, Kahla A, Arunachalam C, Scofield SR, Bowden S, Wallington E, Doohan FM. TaFROG Encodes a Pooideae Orphan Protein That Interacts with SnRK1 and Enhances Resistance to the Mycotoxigenic Fungus Fusarium graminearum. Plant Physiol. 2015 Dec;169(4):2895-906. PMID:26508775 doi:10.1104/pp.15.01056
- ↑ Mariani V, Biasini M, Barbato A, Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013 Nov 1;29(21):2722-8. doi: 10.1093/bioinformatics/btt473., Epub 2013 Aug 27. PMID:23986568 doi:http://dx.doi.org/10.1093/bioinformatics/btt473
- ↑ Chowdhury R, Bouatta N, Biswas S, Floristean C, Kharkar A, Roy K, Rochereau C, Ahdritz G, Zhang J, Church GM, Sorger PK, AlQuraishi M. Single-sequence protein structure prediction using a language model and deep learning. Nat Biotechnol. 2022 Nov;40(11):1617-1623. PMID:36192636 doi:10.1038/s41587-022-01432-w
|