We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Valentina Dutton/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='3mxo' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='3mxo' size='340' side='right' caption='Caption for this structure' scene=''> | ||
This is a default text for your page '''Valentina Dutton/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Valentina Dutton/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | + | ||
== Family == | == Family == | ||
| - | The PGAM5 protein belongs to the phosphoglycerate mutase superfamily. The main characteristic of the proteins in this family is the presence of the PGAM functional domain. However, most of the PGAM proteins are responsible for the transference of the phosphate group in small molecules. The proteins of this family participate in the glycolysis process, more specifically, in the transference of the phosphate group of the compound 3-phosphoglycerate, forming 2-phosphoglycerate | + | The PGAM5 protein belongs to the phosphoglycerate mutase superfamily. The main characteristic of the proteins in this family is the presence of the PGAM functional domain. However, most of the PGAM proteins are responsible for the transference of the phosphate group in small molecules. The proteins of this family participate in the glycolysis process, more specifically, in the transference of the phosphate group of the compound 3-phosphoglycerate, forming 2-phosphoglycerate<ref name="foo">PMID:19590015</ref>. PGAM5 and the T-cell signaling protein suppressor proteins (STS-1 and STS-2) are the only human proteins in this family that have a known phosphatase function<ref name="foo" /><ref name="wil">PMID:25012655</ref>. |
== Structural highlights == | == Structural highlights == | ||
| - | The vast majority of mitochondrial proteins have an N-terminal sequence that indicates that they must be exported to the mitochondria. This sequence is called the mitochondrial signal peptide. When the protein enters the mitochondria, this sequence is usually cleaved, providing greater stability. It is still unknown what this sequence would be in the case of PGAM5 since the protein signal peptide is not cleaved | + | The vast majority of mitochondrial proteins have an N-terminal sequence that indicates that they must be exported to the mitochondria. This sequence is called the mitochondrial signal peptide. When the protein enters the mitochondria, this sequence is usually cleaved, providing greater stability. It is still unknown what this sequence would be in the case of PGAM5 since the protein signal peptide is not cleaved<ref name="sie">PMID:35921890</ref> and PGAM5 is anchored in its entirety to the inner membrane through its transmembrane domain, defined by amino acids 9-29<ref name="cha">PMID:28648608</ref>. |
| - | Regarding the catalytic activity, histidine 105 is responsible for the nucleophilic attack of the phosphate of the target protein, performing the intermediate link between the protein and the phosphate. However, histidine 105 is part of the canonical RHGE motif, present in all proteins of the PGAM family, forming part of the PGAM domain (98-289) | + | Regarding the catalytic activity, histidine 105 is responsible for the nucleophilic attack of the phosphate of the target protein, performing the intermediate link between the protein and the phosphate. However, histidine 105 is part of the canonical RHGE motif, present in all proteins of the PGAM family, forming part of the PGAM domain (98-289)<ref name="cha" />. The WDXNWD motif at amino acids 58-63 has been shown to function as an allosteric regulator of the specific phosphatase activity of PGAM5 by inducing protein oligomerization<ref name="wil" />. Mutations in this motif prevent oligomerization of the enzyme but still present as dimers since the C-terminal tail (270-289) in PGAM5 is responsible for the dimerization of the protein<ref name="cha" />. These dimers, however, do not show phosphatase activity<ref name="wil" />. |
== Function == | == Function == | ||
| - | Even though PGAM5 is a member of the PGAM protein family, it appears to lack phosphoglycerate mutase typical phosphotransferase and/or phosphohydrolase activities. ( | + | Even though PGAM5 is a member of the PGAM protein family, it appears to lack phosphoglycerate mutase typical phosphotransferase and/or phosphohydrolase activities.<ref name="cha" />(UniProt). Instead, this protein is a serine/threonine (Ser/Thr) phosphatase, that is, it’s responsible for protein-protein interactions through dephosphorylation of serine/threonine and, occasionally, histidine residues<ref name="che">PMID:33370650</ref><ref name="shi">PMID:19879837</ref>. Its active site is composed of a histidine residue (His-105) responsible for the nucleophilic attack of the phosphorus atom acting as a phospho-acceptor<ref name="cha" /><ref name="shi" />. |
| - | PGAM5 has been shown to interact with B-cell lymphoma-extra large (Bcl-Xl), an apoptosis regulator, indicating a probable regulating role in the apoptotic process. (Lo and Hannink, 2006 | + | PGAM5 has been shown to interact with B-cell lymphoma-extra large (Bcl-Xl), an apoptosis regulator, indicating a probable regulating role in the apoptotic process.<ref name="foo" /> (Lo and Hannink, 2006). Moreover, it also acts as substrate for both the Kelch-like ECH-associated protein (Keap1) and Nuclear factor erythroid 2-related factor 2 (Nrf2), forming a ternary complex, located in the mitochondria, that regulate gene expression for Nrf2-dependent genes, that are antioxidants and provide protection from reactive oxygen species (ROS) (Lo and Hannink, 2006, 2008). |
Furthermore, it has been studied that cleaved PGAM5 plays an important role in regulation of mitophagy and mitochondrial fission, in cases in which the mitochondria has been damaged. In this instance, PGAM5 is released into the cytosol and can either stay in the cytosol or be translocated to the nucleus (Baba et al, 2021). | Furthermore, it has been studied that cleaved PGAM5 plays an important role in regulation of mitophagy and mitochondrial fission, in cases in which the mitochondria has been damaged. In this instance, PGAM5 is released into the cytosol and can either stay in the cytosol or be translocated to the nucleus (Baba et al, 2021). | ||
| Line 21: | Line 21: | ||
PGAM5 is a mitochondrial enzyme. However, the mitochondria is a complex organelle, composed of really distinct regions. It has 2 membranes, an outer membrane (OMM), in direct contact with the cytoplasm, and an inner membrane (IMM), in contact with the mitochondrial matrix. Between them there is the intermembrane space, where the protons from the matrix are directed Therefore, the mitochondrial matrix exhibits a higher pH than the intermembrane space. | PGAM5 is a mitochondrial enzyme. However, the mitochondria is a complex organelle, composed of really distinct regions. It has 2 membranes, an outer membrane (OMM), in direct contact with the cytoplasm, and an inner membrane (IMM), in contact with the mitochondrial matrix. Between them there is the intermembrane space, where the protons from the matrix are directed Therefore, the mitochondrial matrix exhibits a higher pH than the intermembrane space. | ||
| - | For a long while, PGAM5’s subcellular localization in the mitochondria has been debated. It includes a transmembrane N-terminal domain, which indicates that it would be located in the IMM, facing the intermembrane space (Baba et al., 2021). Nonetheless, it has been demonstrated that PGAM5 interacts with cytosolic proteins, indicating that it could not be located in the IMM, and should be, instead, located in the OMM, facing the outside of the mitochondria | + | For a long while, PGAM5’s subcellular localization in the mitochondria has been debated. It includes a transmembrane N-terminal domain, which indicates that it would be located in the IMM, facing the intermembrane space (Baba et al., 2021). Nonetheless, it has been demonstrated that PGAM5 interacts with cytosolic proteins, indicating that it could not be located in the IMM, and should be, instead, located in the OMM, facing the outside of the mitochondria<ref name="foo" />. More recently, however, studies have shown that the entire protein is exported to the mitochondria, where it’s anchored in the IMM and, during the first stages of mitophagy, it’s cleaved by the PARL protease in the serine 24 residue and it’s then released in the cytosol, where it interacts with other proteins important to this process<ref name="sie" />. |
== Evolutionary conservation == | == Evolutionary conservation == | ||
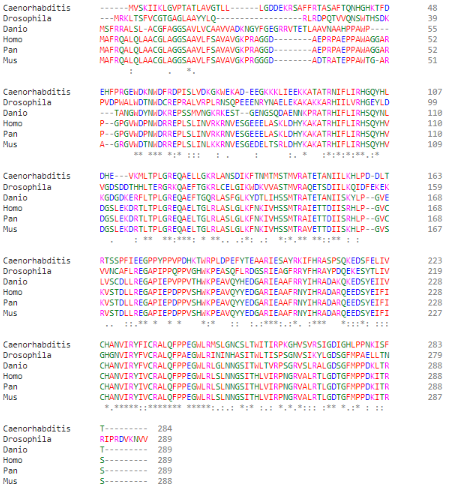
As part of the PGAM protein family, it is expected that PGAM5’s sequence includes a histidine phosphatase superfamily domain typical to this set of proteins, ranging from amino acids 58 up to 287. | As part of the PGAM protein family, it is expected that PGAM5’s sequence includes a histidine phosphatase superfamily domain typical to this set of proteins, ranging from amino acids 58 up to 287. | ||
Revision as of 23:11, 21 June 2023
Human Phosphoglycerate Mutase Family Member 5 (PGAM5)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S, Noguchi T, Kinoshita H, Sekine Y, Iemura S, Natsume T, Ichijo H. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci U S A. 2009 Jul 28;106(30):12301-5. Epub 2009 Jul 9. PMID:19590015 doi:http://dx.doi.org/0901823106
- ↑ 2.0 2.1 2.2 Wilkins JM, McConnell C, Tipton PA, Hannink M. A conserved motif mediates both multimer formation and allosteric activation of phosphoglycerate mutase 5. J Biol Chem. 2014 Sep 5;289(36):25137-48. PMID:25012655 doi:10.1074/jbc.M114.565549
- ↑ 3.0 3.1 Siebert V, Silber M, Heuten E, Muhle-Goll C, Lemberg MK. Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state. J Biol Chem. 2022 Jul 31:102321. doi: 10.1016/j.jbc.2022.102321. PMID:35921890 doi:http://dx.doi.org/10.1016/j.jbc.2022.102321
- ↑ 4.0 4.1 4.2 4.3 4.4 Chaikuad A, Filippakopoulos P, Marcsisin SR, Picaud S, Schroder M, Sekine S, Ichijo H, Engen JR, Takeda K, Knapp S. Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly. Structure. 2017 Jul 5;25(7):1089-1099.e3. doi: 10.1016/j.str.2017.05.020. Epub, 2017 Jun 22. PMID:28648608 doi:http://dx.doi.org/10.1016/j.str.2017.05.020
- ↑ Cheng M, Lin N, Dong D, Ma J, Su J, Sun L. PGAM5: A crucial role in mitochondrial dynamics and programmed cell death. Eur J Cell Biol. 2021 Jan;100(1):151144. PMID:33370650 doi:10.1016/j.ejcb.2020.151144
- ↑ 6.0 6.1 Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009 Oct 30;139(3):468-84. PMID:19879837 doi:10.1016/j.cell.2009.10.006