We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Marcella Maringolo/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
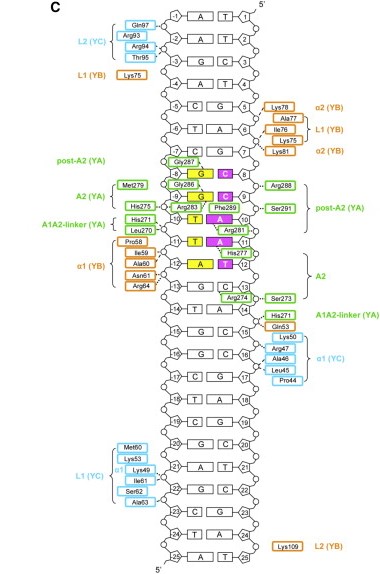
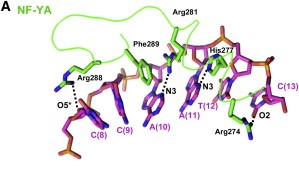
It was hypothesized that NF-YA is recruited by the HFD domain because it would function as a histone tail to allow and stabilize DNA binding with the NF-Y complex. Indeed, the core structures the histone dimer H2A/H2B overlay with NF-YB and NF-YC, while NF-YA overlay with the HB2 tail, indicating a similar function for those structures. | It was hypothesized that NF-YA is recruited by the HFD domain because it would function as a histone tail to allow and stabilize DNA binding with the NF-Y complex. Indeed, the core structures the histone dimer H2A/H2B overlay with NF-YB and NF-YC, while NF-YA overlay with the HB2 tail, indicating a similar function for those structures. | ||
| - | Therefore, the main function of NF-Y is related to its high specificity and affinity binding to DNA, which induces gene promoter activation with the help of other transcriptional factors, although its binding can also induce posttranslational alterations in histones, such as methylations and acetylations, being also capable of establishing acetylation marks directly on gene promoters through recruitment of enzymes responsible for inducing those alterations. This epigenetic marks can induce or repress gene transcription by modifying chromatin accessibility. | + | Therefore, the main function of NF-Y is related to its high specificity and affinity binding to DNA, which induces gene promoter activation with the help of other transcriptional factors, although its binding can also induce posttranslational alterations in histones, such as methylations and acetylations, being also capable of establishing acetylation marks directly on gene promoters through recruitment of enzymes responsible for inducing those alterations. This epigenetic marks can induce or repress gene transcription by modifying chromatin accessibility. In addition, NF-Y can primarily bind into a region where transcription preinitiation complexes are being established, avoiding nucleosome formation and giving space for other TFs to bind to the promoter of a gene that will be transcripted. This primarily binding also can recruit histone modifying enzymes, which regulate chromatin accessibility. |
Here we show the <scene name='97/973093/Human_nf-y/2'>Human NF-Y</scene> molecule structure while binding to the DNA, with each subunit represented in a different color: NF-YA in blue, NF-YB in green and NF-YC in pink, while the DNA is represented in grey. Here is its <scene name='97/973093/Human_nf-y/9'>Backbone</scene> and its <scene name='97/973093/Secondary_structure/1'>Secondary structure</scene> with alpha-helixes colored in pink, and beta-sheets colored in yellow. Using the <scene name='97/973093/Cartoon/1'>cartoon</scene> representation we can see the secondary structures in each subunit. | Here we show the <scene name='97/973093/Human_nf-y/2'>Human NF-Y</scene> molecule structure while binding to the DNA, with each subunit represented in a different color: NF-YA in blue, NF-YB in green and NF-YC in pink, while the DNA is represented in grey. Here is its <scene name='97/973093/Human_nf-y/9'>Backbone</scene> and its <scene name='97/973093/Secondary_structure/1'>Secondary structure</scene> with alpha-helixes colored in pink, and beta-sheets colored in yellow. Using the <scene name='97/973093/Cartoon/1'>cartoon</scene> representation we can see the secondary structures in each subunit. | ||
Revision as of 12:53, 23 June 2023
| |||||||||||