User:Marcella Maringolo/Sandbox 1
From Proteopedia
| Line 10: | Line 10: | ||
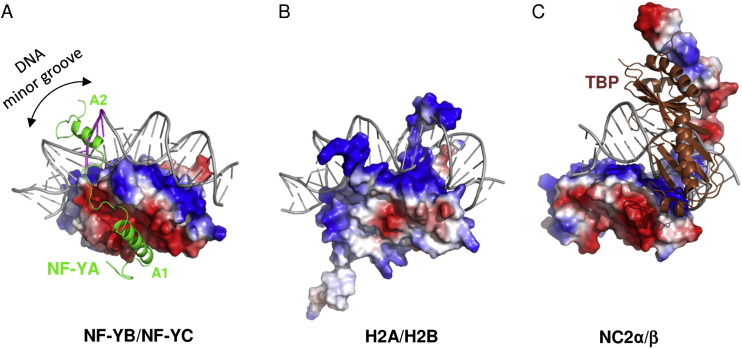
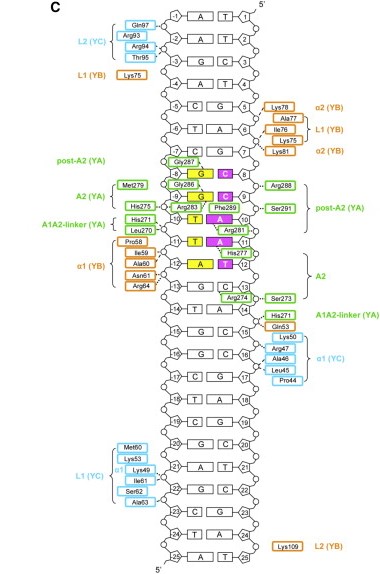
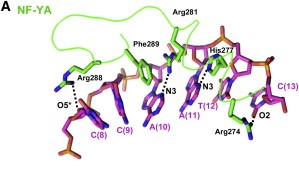
The key structural feature of the NF-Y/DNA complex is the minor-groove interaction of NF-YA, which induces an approximately 80° bend in the DNA. The structure and DNA-binding mode of NF-YB/NF-YC H FDs are similar to those of the core histones H2A/H2B, TATA-binding protein (TBP)-associated factors (TAFs), the TBP/ TATA-binding negative cofactor 2 (NC2α/β), and the CHRAC15/CHRAC17 subunits of the nucleosome remodeling complex CHRAC. Yet, unlike H2A/H2B which lack sequence specificity, NF-YB/NF-YC interaction with NF-YA provides the NF-Y complex with sequence-specific targeting capability as well as nucleosome-like properties of non-specific DNA binding, a combination that allows for stable DNA binding. NF-Y, largely described as a transcription activator via its promoter-proximal binding, is a key regulator of cell cycle progression in proliferating cells, with its activity often downregulated during cellular differentiation and senescence. In addition to binding core promoters, NF-Y has also been shown to bind enhancer elements away from TSSs, but its function and mechanism of action at these distal regulatory elements remain to be elucidated | The key structural feature of the NF-Y/DNA complex is the minor-groove interaction of NF-YA, which induces an approximately 80° bend in the DNA. The structure and DNA-binding mode of NF-YB/NF-YC H FDs are similar to those of the core histones H2A/H2B, TATA-binding protein (TBP)-associated factors (TAFs), the TBP/ TATA-binding negative cofactor 2 (NC2α/β), and the CHRAC15/CHRAC17 subunits of the nucleosome remodeling complex CHRAC. Yet, unlike H2A/H2B which lack sequence specificity, NF-YB/NF-YC interaction with NF-YA provides the NF-Y complex with sequence-specific targeting capability as well as nucleosome-like properties of non-specific DNA binding, a combination that allows for stable DNA binding. NF-Y, largely described as a transcription activator via its promoter-proximal binding, is a key regulator of cell cycle progression in proliferating cells, with its activity often downregulated during cellular differentiation and senescence. In addition to binding core promoters, NF-Y has also been shown to bind enhancer elements away from TSSs, but its function and mechanism of action at these distal regulatory elements remain to be elucidated | ||
| + | |||
| + | [[Image: Diferences.jpg]] | ||
== Function and Structural highlights == | == Function and Structural highlights == | ||
| Line 27: | Line 29: | ||
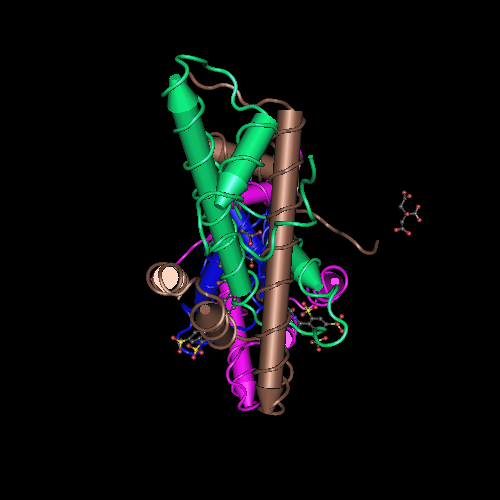
Here we show the <scene name='97/973093/Human_nf-y/2'>Human NF-Y</scene> molecule structure while binding to the DNA, with each subunit represented in a different color: NF-YA in blue, NF-YB in green and NF-YC in pink, while the DNA is represented in grey. Here is its <scene name='97/973093/Human_nf-y/9'>Backbone</scene> and its <scene name='97/973093/Secondary_structure/1'>Secondary structure</scene> with alpha-helixes colored in pink, and beta-sheets colored in yellow. Using the <scene name='97/973093/Cartoon/1'>cartoon</scene> representation we can see the secondary structures in each subunit. | Here we show the <scene name='97/973093/Human_nf-y/2'>Human NF-Y</scene> molecule structure while binding to the DNA, with each subunit represented in a different color: NF-YA in blue, NF-YB in green and NF-YC in pink, while the DNA is represented in grey. Here is its <scene name='97/973093/Human_nf-y/9'>Backbone</scene> and its <scene name='97/973093/Secondary_structure/1'>Secondary structure</scene> with alpha-helixes colored in pink, and beta-sheets colored in yellow. Using the <scene name='97/973093/Cartoon/1'>cartoon</scene> representation we can see the secondary structures in each subunit. | ||
| - | + | ||
==NF-Y and diseases== | ==NF-Y and diseases== | ||
| Line 36: | Line 38: | ||
Other circumstances where mutations of the NF-Y binding site occur can result in the block of NF-Y binding, altering the expression of genes regulated by NF-Y transcriptional regulation. | Other circumstances where mutations of the NF-Y binding site occur can result in the block of NF-Y binding, altering the expression of genes regulated by NF-Y transcriptional regulation. | ||
| + | Currently there are many studies about the inhibition of NF-Y as a stabilizer of many tumors, since the complex is found in large amounts in cancer cells and allows their proliferation. Suramin is one of the most used inhibitors, where it prevents NF-Y and DNA binding | ||
| + | |||
| + | [[Image: Mmdbimage.png]] | ||
Revision as of 22:07, 24 June 2023
| |||||||||||
References
Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012 Jan-Feb;47(1):29-49. doi: 10.3109/10409238.2011.628970.
McNabb DS, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes & Development. 1995 Jan;9(1):47-58. DOI: 10.1101/gad.9.1.47. PMID: 7828851.
Nardini, Marco, et al. “Sequence-Specific Transcription Factor NF-Y Displays Histone-like DNA Binding and H2B-like Ubiquitination.” Cell, vol. 152, no. 1-2, 17 Jan. 2013, pp. 132–143, https://doi.org/10.1016/j.cell.2012.11.047.
Oldfield AJ, Henriques T, Kumar D, Burkholder AB, Cinghu S, Paulet D, Bennett BD, Yang P, Scruggs BS, Lavender CA, Rivals E, Adelman K, Jothi R. NF-Y controls fidelity of transcription initiation at gene promoters through maintenance of the nucleosome-depleted region. Nat Commun. 2019 Jul 11;10(1):3072. doi: 10.1038/s41467-019-10905-7.