We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Porin
From Proteopedia
(Difference between revisions)
| Line 21: | Line 21: | ||
== Structure == | == Structure == | ||
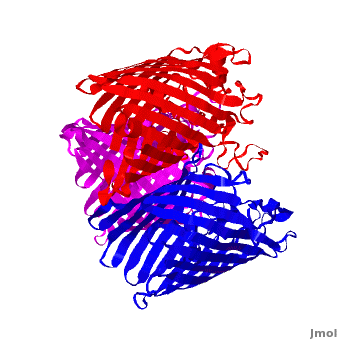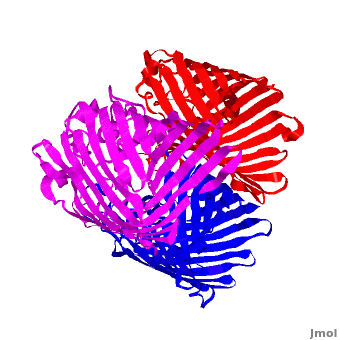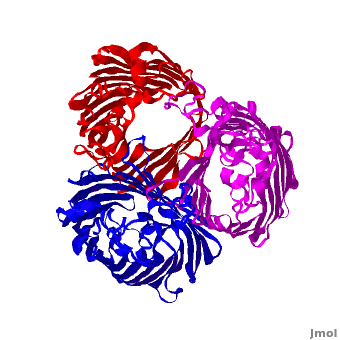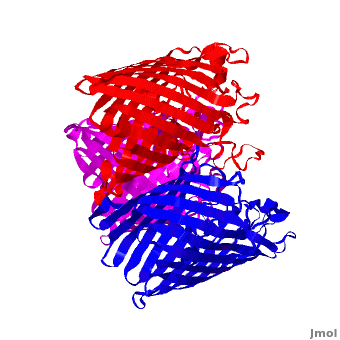
| - | One representative porin structure is the crystal structure osmoporin OmpC from ''Escherichia coli'' ([[2j1n]]). OmpC has three beta-barrels associated to form a <scene name='Porin/Cv/2'>tight trimer</scene> <ref>PMID:16949612</ref>. Porin is a transmembrane protein, as can be seen from the <jmol><jmolLink><script>script "/scripts/1a0s/Hidrophobic/1.spt"; ppdiaCaptionCmd = "changeCaption('Hydrophobic residues (shown in tan) are prevalent where the protein comes in contact with the | + | One representative porin structure is the crystal structure osmoporin OmpC from ''Escherichia coli'' ([[2j1n]]). OmpC has three beta-barrels associated to form a <scene name='Porin/Cv/2'>tight trimer</scene> <ref>PMID:16949612</ref>. |
| + | |||
| + | Porin is a <scene name='41/411405/Membrane/1'>transmembrane protein</scene>, as can be seen from the <jmol><jmolLink><script>script "/scripts/1a0s/Hidrophobic/1.spt"; ppdiaCaptionCmd = "changeCaption('Hydrophobic residues (shown in tan) are prevalent where the protein comes in contact with the hydrophobic layer of the double-layer membrane, while other parts of the surface are hydrophilic (hydrophilic residues, ordered water molecules and calcium ions shown in skyblue). Shown here is the sucrose-specific porin (PDB-ID 1a0s) in its trimeric quaternary structure.','white','black');";javascript @ppdiaCaptionCmd;</script><text>hydrophobic ring</text></jmolLink></jmol> around the protein, this makes it possible to submerge in the lipid bilayer (hydrophobic amino acids are sandybrown, hydrophilic ones are cyan). As you can <scene name='1a0s/Hidrophobic1/1'>see</scene> the channel in the protein is made of mainly hydrophilic chains thus making it possible for the sugar to pass through (these scenes were created by Nádori Gergely). | ||
The channels have wide openings on either side with a tighter bottleneck in the middle, as illustrated in the interactive view <scene name='99/995028/Pacupp/2'>visualizing channels</scene> with pseudoatoms. In an alternative visualization, channels are shown as <scene name='41/411405/Channels/3'>surfaces</scene>, slabbed on both sides of the bottleneck for better visibility. | The channels have wide openings on either side with a tighter bottleneck in the middle, as illustrated in the interactive view <scene name='99/995028/Pacupp/2'>visualizing channels</scene> with pseudoatoms. In an alternative visualization, channels are shown as <scene name='41/411405/Channels/3'>surfaces</scene>, slabbed on both sides of the bottleneck for better visibility. | ||
Revision as of 14:14, 19 September 2023
| |||||||||||
Acknowledgement
The scene showing channels as pseudoatoms is from a page (User:Eric_Martz/Sandbox_19) made by Eric Martz. Eric also helped creating the surface rendition of the channels (technical details here: Image:Tunnels.jvxl).
References
- ↑ Shoshan-Barmatz V, Israelson A, Brdiczka D, Sheu SS. The voltage-dependent anion channel (VDAC): function in intracellular signalling, cell life and cell death. Curr Pharm Des. 2006;12(18):2249-70. PMID:16787253
- ↑ Van Gelder P, Dumas F, Bartoldus I, Saint N, Prilipov A, Winterhalter M, Wang Y, Philippsen A, Rosenbusch JP, Schirmer T. Sugar transport through maltoporin of Escherichia coli: role of the greasy slide. J Bacteriol. 2002 Jun;184(11):2994-9. PMID:12003940
- ↑ Suginta W, Chumjan W, Mahendran KR, Schulte A, Winterhalter M. Chitoporin from Vibrio harveyi, a channel with exceptional sugar specificity. J Biol Chem. 2013 Apr 19;288(16):11038-46. PMID:23447539 doi:10.1074/jbc.M113.454108
- ↑ Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J Mol Biol. 2006 Oct 6;362(5):933-42. Epub 2006 Aug 3. PMID:16949612 doi:10.1016/j.jmb.2006.08.002
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Karsten Theis, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky