We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Ben Whiteside
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
The <scene name='10/1038819/Amidated_c_term/9'>C-Terminus</scene> of amylin contains an amide group, rather than a carboxylic acid group. This chemical modification allows for more extensive hydrogen bonding to nearby residues, due to the added hydrogen bond donor on the NH2 group. In turn, this allows for favorable hydrogen bonds between S129 of the transmembrane domain and the main chain of Y37 on amylin. This interaction causes a "kink" in the random coil of amylin, displacing Y37 into a hydrophobic pocket, allowing for favorable hydrophobic interactions with W79 of the transmembrane domain. This amidation is thought to be a post-translational modification. | The <scene name='10/1038819/Amidated_c_term/9'>C-Terminus</scene> of amylin contains an amide group, rather than a carboxylic acid group. This chemical modification allows for more extensive hydrogen bonding to nearby residues, due to the added hydrogen bond donor on the NH2 group. In turn, this allows for favorable hydrogen bonds between S129 of the transmembrane domain and the main chain of Y37 on amylin. This interaction causes a "kink" in the random coil of amylin, displacing Y37 into a hydrophobic pocket, allowing for favorable hydrophobic interactions with W79 of the transmembrane domain. This amidation is thought to be a post-translational modification. | ||
=== Two-Domain Model of Amylin Binding === | === Two-Domain Model of Amylin Binding === | ||
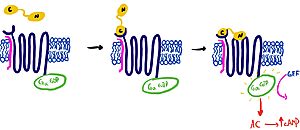
| - | It is hypothesized that amylin binds to the receptor via a two-domain model. The model suggests a series of steps for how amylin binds. First, the c-terminus of amylin binds to the n terminus of the extracellular domain of the receptor. This binding factors the alignment of amylin's n-terminus to the primary GPCR binding site. This activates the GPCR, leading to subsequent activation of adenylyl cyclase and cAMP release. [[Image:Domain_drawingnew.jpg|300px|left|thumb|Figure | + | It is hypothesized that amylin binds to the receptor via a two-domain model. The model suggests a series of steps for how amylin binds. First, the c-terminus of amylin binds to the n terminus of the extracellular domain of the receptor. This binding factors the alignment of amylin's n-terminus to the primary GPCR binding site. This activates the GPCR, leading to subsequent activation of adenylyl cyclase and cAMP release. [[Image:Domain_drawingnew.jpg|300px|left|thumb|Figure 1: The Two Domain Model]] |
===RAMP-CTR Interface=== | ===RAMP-CTR Interface=== | ||
<scene name='10/1038828/Ramp_ctr_interface/9'>RAMP CTR Interface </scene> is a key interaction that stabilizes the protein complex and positions the receptor to favorably bind to amylin. The RAMP-CTR interface extends into the plasma membrane, providing additional non-covalent bonding between the protein complex and the cell membrane. | <scene name='10/1038828/Ramp_ctr_interface/9'>RAMP CTR Interface </scene> is a key interaction that stabilizes the protein complex and positions the receptor to favorably bind to amylin. The RAMP-CTR interface extends into the plasma membrane, providing additional non-covalent bonding between the protein complex and the cell membrane. | ||
| Line 30: | Line 30: | ||
It has been thought that [https://en.wikipedia.org/wiki/Missense_mutation missense mutations] in residues C2 and C7 of the amylin peptide could lead to an increased risk of Alzheimer's Disease (CITE) <ref name=”Grizzanti”>PMID: 30282360</ref>. Because of the rigidity these cysteine resides provide, reductions of their disulfide interaction leads to an increased risk of amyloid plaques due to amylin misfolding and forming aggregates. During drug design, pharmaceutical companies have focused on maintaining amylin residues C2 and C7, as well as K1, which forms a hydrogen bond donor for the <scene name='10/1038828/N_term_disulfidenew/1'>E294 Side Chain</scene> and main chain carbonyl. Additionally, pharmaceuticals companies have also opted to maintain residues <scene name='10/1038819/Amidated_c_term/9'>Y37 and T36</scene>, which are critical residues in stabilizing the C terminus of amylin to the receptor binding site. While there are hardly any differences in the helical portion of amylin and the synthetic analogue pramlintide, there is a difference in the extended random coil at the C terminus. | It has been thought that [https://en.wikipedia.org/wiki/Missense_mutation missense mutations] in residues C2 and C7 of the amylin peptide could lead to an increased risk of Alzheimer's Disease (CITE) <ref name=”Grizzanti”>PMID: 30282360</ref>. Because of the rigidity these cysteine resides provide, reductions of their disulfide interaction leads to an increased risk of amyloid plaques due to amylin misfolding and forming aggregates. During drug design, pharmaceutical companies have focused on maintaining amylin residues C2 and C7, as well as K1, which forms a hydrogen bond donor for the <scene name='10/1038828/N_term_disulfidenew/1'>E294 Side Chain</scene> and main chain carbonyl. Additionally, pharmaceuticals companies have also opted to maintain residues <scene name='10/1038819/Amidated_c_term/9'>Y37 and T36</scene>, which are critical residues in stabilizing the C terminus of amylin to the receptor binding site. While there are hardly any differences in the helical portion of amylin and the synthetic analogue pramlintide, there is a difference in the extended random coil at the C terminus. | ||
| - | In order to restore basal amylin levels in mice, researchers have performed PEGylation, the addition of poly ethylene glycol, to amylin within residues 1-11 of the peptide, most likely at the two amine groups of K1<ref name=”Guerreiro” | + | In order to restore basal amylin levels in mice, researchers have performed PEGylation, the addition of poly ethylene glycol, to amylin within residues 1-11 of the peptide, most likely at the two amine groups of K1<ref name=”Guerreiro”>. Administration of the modified amylin in mice showed evidence of reduced glycemia and prolonged action compared to endogenous amylin<ref name=”Guerreiro”>PMID: 23818080</ref>. |
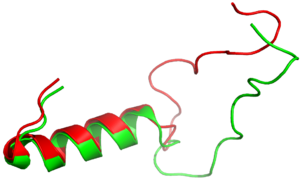
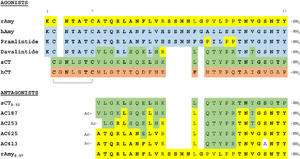
| - | [[Image:align.png|300px|left|thumb|Figure | + | [[Image:align.png|300px|left|thumb|Figure 2:Amylin (green) aligned with Pramlintide (red)]] [[Image:pram sequence align.png|300px|left|thumb|Figure 3:Pramlintide Sequence alignment with varying forms of amylin. Atoms C2—C7 and K1 of the N-terminal region are conserved. Y37 and T36 of the C-terminal region are also conserved.]] |
Revision as of 13:50, 25 April 2024
AMYR
| |||||||||||
References
- ↑ Bower RL, Hay DL. Amylin structure-function relationships and receptor pharmacology: implications for amylin mimetic drug development. Br J Pharmacol. 2016 Jun;173(12):1883-98. PMID:27061187 doi:10.1111/bph.13496
- ↑ Hay DL, Chen S, Lutz TA, Parkes DG, Roth JD. Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol Rev. 2015 Jul;67(3):564-600. PMID:26071095 doi:10.1124/pr.115.010629
- ↑ Cao J, Belousoff MJ, Liang YL, Johnson RM, Josephs TM, Fletcher MM, Christopoulos A, Hay DL, Danev R, Wootten D, Sexton PM. A structural basis for amylin receptor phenotype. Science. 2022 Mar 25;375(6587):eabm9609. PMID:35324283 doi:10.1126/science.abm9609
- ↑ Parameswaran N, Spielman WS. RAMPs: The past, present and future. Trends Biochem Sci. 2006 Nov;31(11):631-8. PMID:17010614 doi:10.1016/j.tibs.2006.09.006
- ↑ Hay DL, Pioszak AA. Receptor Activity-Modifying Proteins (RAMPs): New Insights and Roles. Annu Rev Pharmacol Toxicol. 2016;56:469-87. PMID:26514202 doi:10.1146/annurev-pharmtox-010715-103120
- ↑ Hay DL, Chen S, Lutz TA, Parkes DG, Roth JD. Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol Rev. 2015 Jul;67(3):564-600. PMID:26071095 doi:10.1124/pr.115.010629
- ↑ Hoogwerf BJ, Doshi KB, Diab D. Pramlintide, the synthetic analogue of amylin: physiology, pathophysiology, and effects on glycemic control, body weight, and selected biomarkers of vascular risk. Vasc Health Risk Manag. 2008;4(2):355-62. PMID:18561511 doi:10.2147/vhrm.s1978
- ↑ Gingell JJ, Burns ER, Hay DL. Activity of pramlintide, rat and human amylin but not Aβ1-42 at human amylin receptors. Endocrinology. 2014 Jan;155(1):21-6. PMID:24169554 doi:10.1210/en.2013-1658
- ↑ Grizzanti J, Corrigan R, Casadesus G. Neuroprotective Effects of Amylin Analogues on Alzheimer's Disease Pathogenesis and Cognition. J Alzheimers Dis. 2018;66(1):11-23. PMID:30282360 doi:10.3233/JAD-180433
- ↑ . Administration of the modified amylin in mice showed evidence of reduced glycemia and prolonged action compared to endogenous amylin<ref>PMID: 23818080</li></ol></ref>
Student Contributors
Andrew Helmerich, Mathias Vander Eide, Ben Whiteside