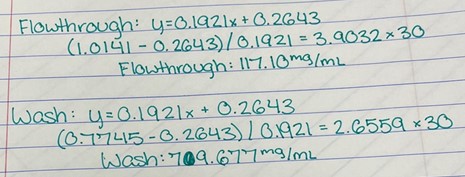We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox326
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== '''Introduction''' == | == '''Introduction''' == | ||
| - | == Research Question == | + | === Research Question === |
What is the function and qualities (location and optimal conditions) of our assigned unknown protein, 2QRU? | What is the function and qualities (location and optimal conditions) of our assigned unknown protein, 2QRU? | ||
| - | == Problem Relevance == | + | === Problem Relevance === |
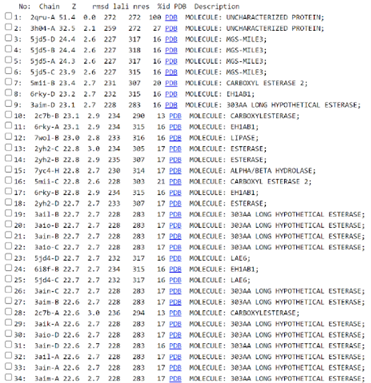
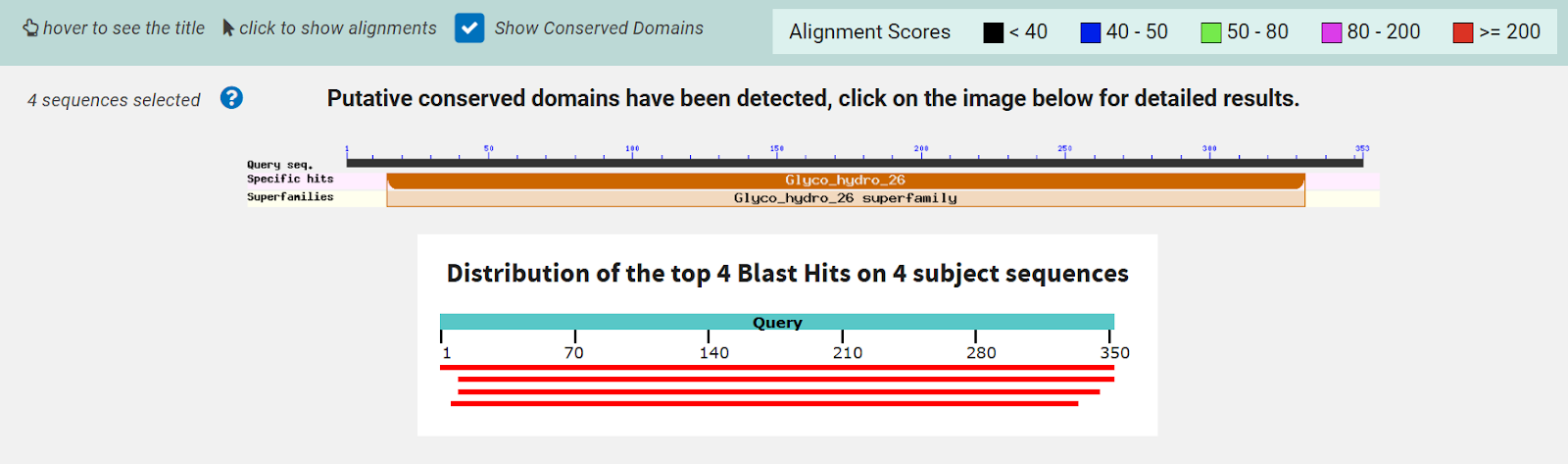
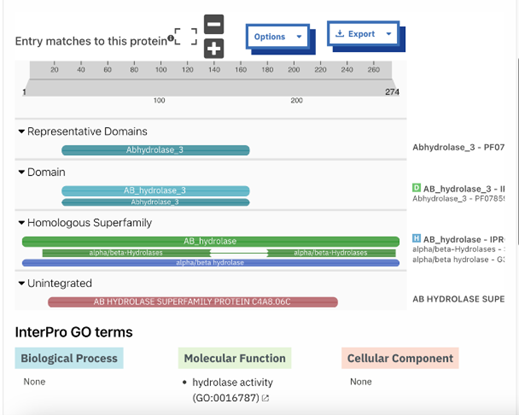
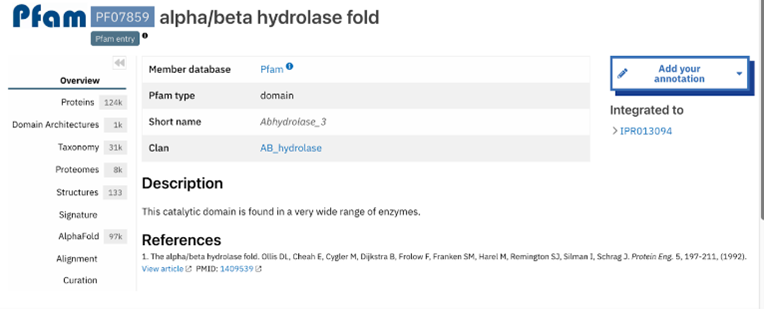
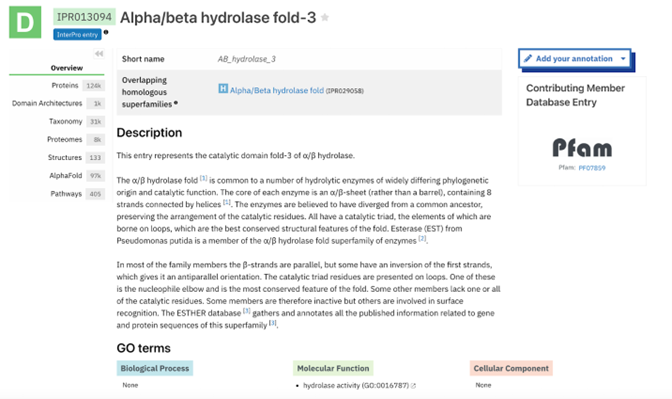
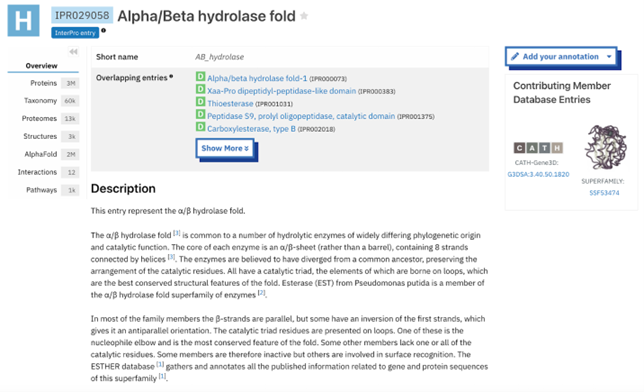
The function of this protein is not known past the fact that it is in the super family alpha/beta hydrolase. Knowing the function of this protein could be helpful since knowing a protein’s function allows for extensive classification of said protein. | The function of this protein is not known past the fact that it is in the super family alpha/beta hydrolase. Knowing the function of this protein could be helpful since knowing a protein’s function allows for extensive classification of said protein. | ||
| - | == Research Significance == | + | === Research Significance === |
Identifying the structure of the unknown protein would reveal its function which could be used in many different ways. By connecting the protein’s function to symptoms and mechanisms, it could be used as a target for treatment of various diseases. | Identifying the structure of the unknown protein would reveal its function which could be used in many different ways. By connecting the protein’s function to symptoms and mechanisms, it could be used as a target for treatment of various diseases. | ||
| - | == Hypothesis == | + | === Hypothesis === |
Based on the super family classification, protein 2QRU is a type of hydrolase. Running experiments to test the protein under three different pH conditions based on the three most common locations of hydrolases (digestive system, neurons, and muscle cells) will reveal the activity of the protein in these different pH’s and therefore could suggest where the protein is found and consequently its function. | Based on the super family classification, protein 2QRU is a type of hydrolase. Running experiments to test the protein under three different pH conditions based on the three most common locations of hydrolases (digestive system, neurons, and muscle cells) will reveal the activity of the protein in these different pH’s and therefore could suggest where the protein is found and consequently its function. | ||
| - | == How Experimental Data Will Answer the Hypothesis == | + | === How Experimental Data Will Answer the Hypothesis === |
* The different computer modeling strategies used such as Chimera, SPRITE, Dali, BLAST, InterPRO, and SwissDock allowed visualization of the protein’s structure as a whole | * The different computer modeling strategies used such as Chimera, SPRITE, Dali, BLAST, InterPRO, and SwissDock allowed visualization of the protein’s structure as a whole | ||
| Line 40: | Line 40: | ||
== '''Materials & Methods''' == | == '''Materials & Methods''' == | ||
| - | == Computer Modeling Strategies == | + | === Computer Modeling Strategies === |
* Chimera: allowed visualization of the protein; was used to compare conformation, identity, and conservation of amino acid side chains; was used instead of Dali (Dali didn’t analyze side chains); visualized results from SPRITE in a different way. | * Chimera: allowed visualization of the protein; was used to compare conformation, identity, and conservation of amino acid side chains; was used instead of Dali (Dali didn’t analyze side chains); visualized results from SPRITE in a different way. | ||
| Line 55: | Line 55: | ||
| - | == Key Information about Chemicals, Equipment, & Instrumentation == | + | === Key Information about Chemicals, Equipment, & Instrumentation === |
| - | * Escherichia coli (E. coli) strain | + | * Escherichia coli (E. coli) strain BL21(DE3) |
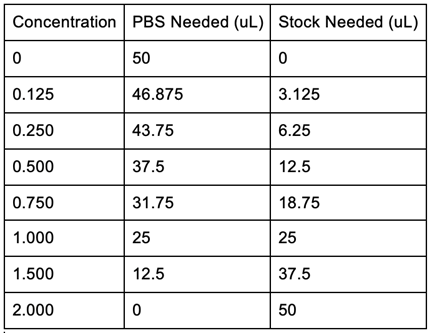
* Buffers (Sodium Phosphate buffer, Tris-HCl, Re-Suspension Buffer, Cell Lysis Buffer, 1X Wash Buffer, 1X Elution Buffer, 10X SDS-PAGE Buffer, Coomassie Blue Stain, and Destain) were prepared by classmates with appropriate techniques and all materials were stored at necessary temperatures. | * Buffers (Sodium Phosphate buffer, Tris-HCl, Re-Suspension Buffer, Cell Lysis Buffer, 1X Wash Buffer, 1X Elution Buffer, 10X SDS-PAGE Buffer, Coomassie Blue Stain, and Destain) were prepared by classmates with appropriate techniques and all materials were stored at necessary temperatures. | ||
| Line 200: | Line 200: | ||
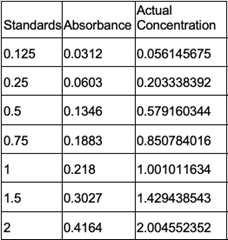
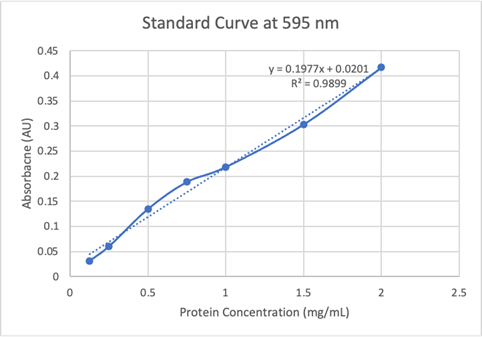
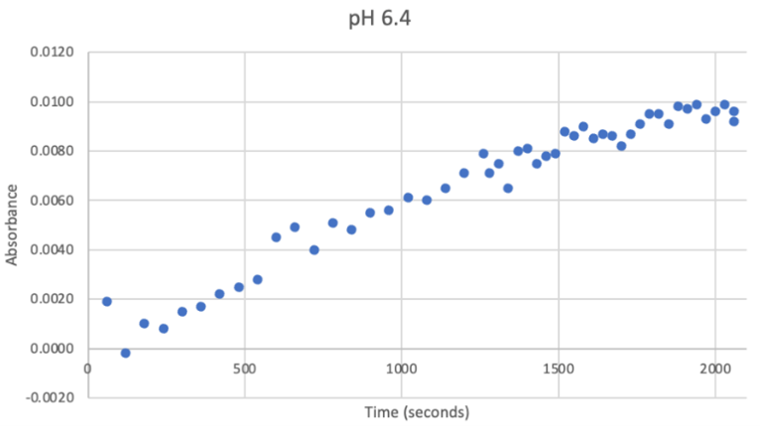
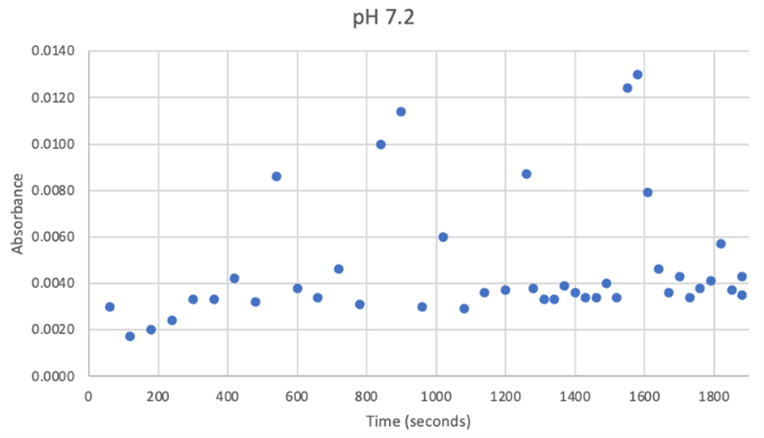
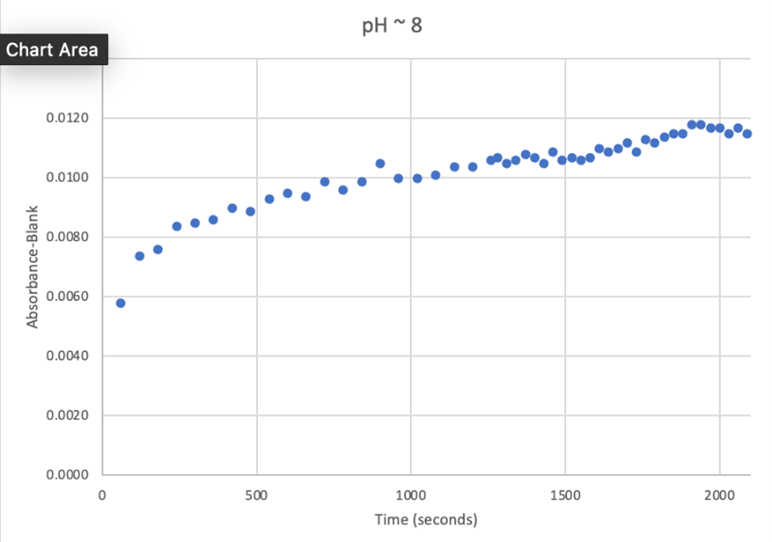
Since the unknown enzyme had a recognizable activity with the common hydrolase substrate, p-nitrophenyl acetate, it is likely that 2QRU is indeed a hydrolase. When unknown protein 2QRU was tested in a pH of ~2, the enzyme completely dissociated from solution indicating it can in no way function in that acidic of an environment in the body. Since the protein worked best in the pH around neutral, it is likely that the enzyme functions as a hydrolase in either neuronal or muscle cells. | Since the unknown enzyme had a recognizable activity with the common hydrolase substrate, p-nitrophenyl acetate, it is likely that 2QRU is indeed a hydrolase. When unknown protein 2QRU was tested in a pH of ~2, the enzyme completely dissociated from solution indicating it can in no way function in that acidic of an environment in the body. Since the protein worked best in the pH around neutral, it is likely that the enzyme functions as a hydrolase in either neuronal or muscle cells. | ||
| - | == Accuracy & Precision of Results == | + | === Accuracy & Precision of Results === |
Our results are partially accurate as we tested our protein with a general substrate for hydrolases. Although not directly associated with our enzymes exact active site, the substrate we used, p-nitrophenyl acetate (PNPA), does generally work with hydrolases as a whole. In terms of our results from the computer models and directly from the equipment used, those results are accurate as the databases are reliable and the equipment was calibrated and used correctly. | Our results are partially accurate as we tested our protein with a general substrate for hydrolases. Although not directly associated with our enzymes exact active site, the substrate we used, p-nitrophenyl acetate (PNPA), does generally work with hydrolases as a whole. In terms of our results from the computer models and directly from the equipment used, those results are accurate as the databases are reliable and the equipment was calibrated and used correctly. | ||
| Line 206: | Line 206: | ||
Our results may be precise, but we would need further testing and repeats of our results to determine whether they are statistically different or precise enough to be the same. We only ran our experiments and each of our pH levels once, so we cannot confidently confirm precision. | Our results may be precise, but we would need further testing and repeats of our results to determine whether they are statistically different or precise enough to be the same. We only ran our experiments and each of our pH levels once, so we cannot confidently confirm precision. | ||
| - | == Future Experiments == | + | === Future Experiments === |
Using a substrate that has been shown in the computer modeling to work more directly with the expected active site of our protein can give more specific insight into the type of hydrolase that 2QRU may be. | Using a substrate that has been shown in the computer modeling to work more directly with the expected active site of our protein can give more specific insight into the type of hydrolase that 2QRU may be. | ||
Revision as of 19:52, 26 April 2024
Identification of Unknown Protein 2QRU
| |||||||||||