We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox324
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== Overview of Esterase == | == Overview of Esterase == | ||
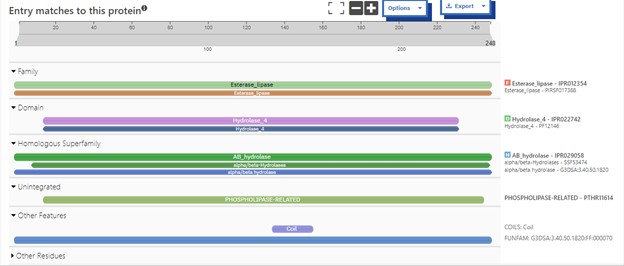
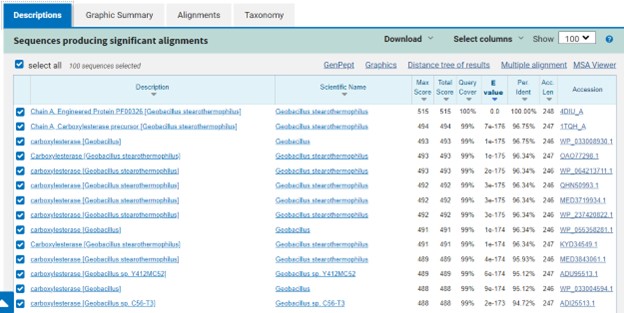
| - | This protein belongs to the esterase family and expresses alpha/beta-hydrolase activity. Esterases are broadly defined as hydrolases that split ester, amide, and/or thioester bonds in certain compounds that cause prodrug activation and/or detoxification. The ability of esterases to hydrolyze various drugs make them an important component in drug metabolism<ref>Fukami, T.; Yokoi, T. The Emerging Role of Human Esterases. Drug Metabolism and Pharmacokinetics 2012, 27 (5), 466–477. https://doi.org/10.2133/dmpk.dmpk-12-rv-042.</ref>. This protein's close association with carboxylesterases also indicates that this protein is potentially involved in the hydrolysis of carboxylic ester bonds. Drug metabolism (drug hydrolysis, drug absorption<ref>Williams, F. M. Clinical Significance of Esterases in Man. Clinical pharmacokinetics 1985, 10 (5), 392–403. https://doi.org/10.2165/00003088-198510050-00002.</ref>, etc.) would be inhibited if the function of an esterase is disrupted. | + | This protein belongs to the esterase family and expresses alpha/beta-hydrolase activity. Esterases are broadly defined as hydrolases that split ester, amide, and/or thioester bonds in certain compounds that cause prodrug activation and/or detoxification. The ability of esterases to hydrolyze various drugs make them an important component in drug metabolism<ref>Fukami, T.; Yokoi, T. The Emerging Role of Human Esterases. Drug Metabolism and Pharmacokinetics 2012, 27 (5), 466–477. https://doi.org/10.2133/dmpk.dmpk-12-rv-042.</ref>. This protein's close association with carboxylesterases also indicates that this protein is potentially involved in the hydrolysis of carboxylic ester bonds. Esterases are most commonly located in the skin and liver of mammals, with carboxylesterases being frequently found in the cytosol and endoplasmic reticulum of the skin<ref>Tokudome, Y.; Katayanagi, M.; Hashimoto, F. Esterase Activity and Intracellular Localization in Reconstructed Human Epidermal Cultured Skin Models. Annals of Dermatology 2015, 27 (3), 269. https://doi.org/10.5021/ad.2015.27.3.269.</ref>. Drug metabolism (drug hydrolysis, drug absorption<ref>Williams, F. M. Clinical Significance of Esterases in Man. Clinical pharmacokinetics 1985, 10 (5), 392–403. https://doi.org/10.2165/00003088-198510050-00002.</ref>, etc.) would be inhibited if the function of an esterase is disrupted. |
== Proposed Function == | == Proposed Function == | ||
| Line 50: | Line 50: | ||
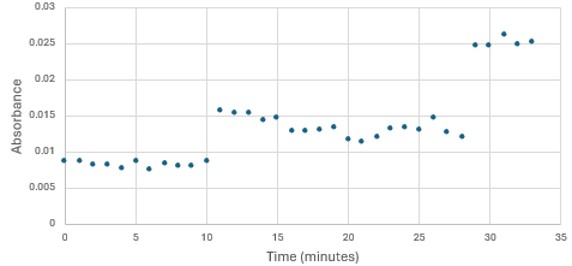
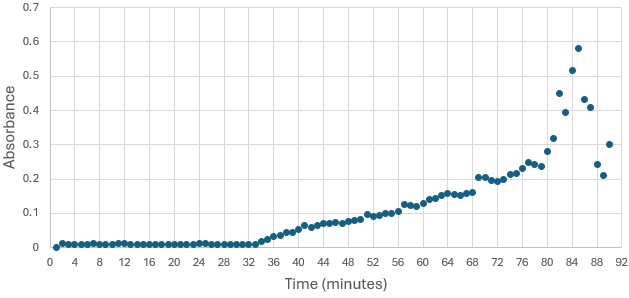
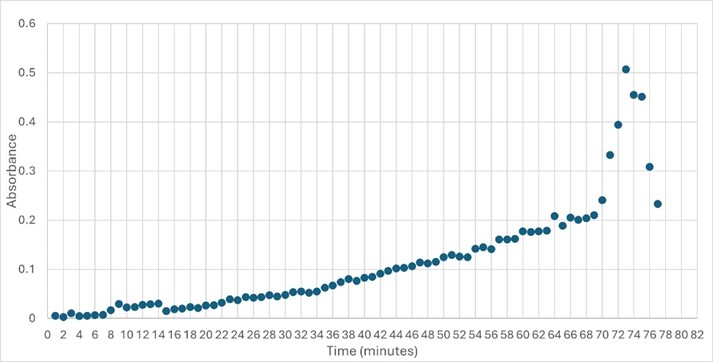
Highlight the data that helped you come to your conclusion here including any relevant figures. Make sure to include potential substrates and binding sites. | Highlight the data that helped you come to your conclusion here including any relevant figures. Make sure to include potential substrates and binding sites. | ||
| + | |||
| + | [Write about potential substrates] | ||
== Conclusions == | == Conclusions == | ||
Revision as of 14:41, 29 April 2024
| |||||||||||
References
- ↑ Fukami, T.; Yokoi, T. The Emerging Role of Human Esterases. Drug Metabolism and Pharmacokinetics 2012, 27 (5), 466–477. https://doi.org/10.2133/dmpk.dmpk-12-rv-042.
- ↑ Tokudome, Y.; Katayanagi, M.; Hashimoto, F. Esterase Activity and Intracellular Localization in Reconstructed Human Epidermal Cultured Skin Models. Annals of Dermatology 2015, 27 (3), 269. https://doi.org/10.5021/ad.2015.27.3.269.
- ↑ Williams, F. M. Clinical Significance of Esterases in Man. Clinical pharmacokinetics 1985, 10 (5), 392–403. https://doi.org/10.2165/00003088-198510050-00002.
- ↑ Zhang, S.; Sun, W.; Xu, L.; Zheng, X.; Chu, X.; Tian, J.; Wu, N.; Fan, Y. Identification of the Para-Nitrophenol Catabolic Pathway, and Characterization of Three Enzymes Involved in the Hydroquinone Pathway, in Pseudomonas Sp. 1-7. BMC Microbiology 2012, 12 (1). https://doi.org/10.1186/1471-2180-12-27.
- ↑ Vázquez-Mayorga, E.; Díaz-Sánchez, Á.; Dagda, R.; Domínguez-Solís, C.; Dagda, R.; Coronado-Ramírez, C.; Martínez-Martínez, A. Novel Redox-Dependent Esterase Activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson’s Disease. International Journal of Molecular Sciences 2016, 17 (8), 1346. https://doi.org/10.3390/ijms17081346.
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644