We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox324
From Proteopedia
(Difference between revisions)
| Line 46: | Line 46: | ||
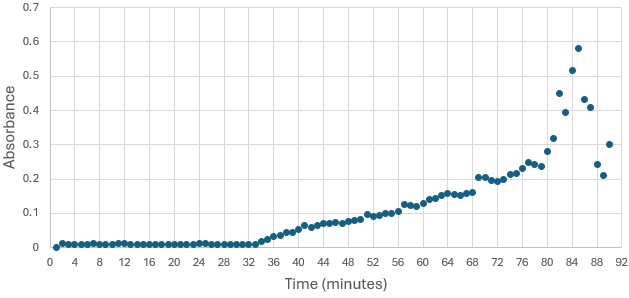
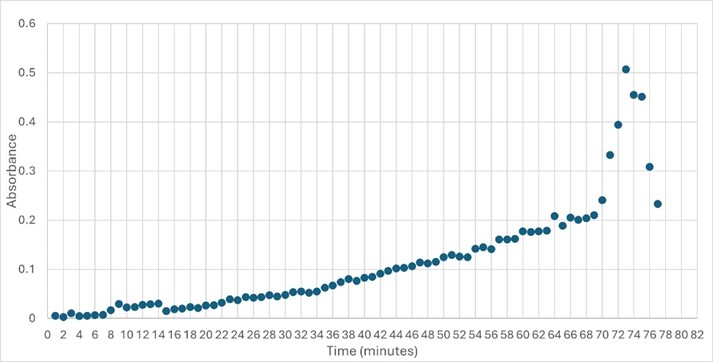
'''Figure 5: ''' Trial 2 of 4DIU enzymatic activity measured over time via change in absorbance at 405nm in pH 6 buffer | '''Figure 5: ''' Trial 2 of 4DIU enzymatic activity measured over time via change in absorbance at 405nm in pH 6 buffer | ||
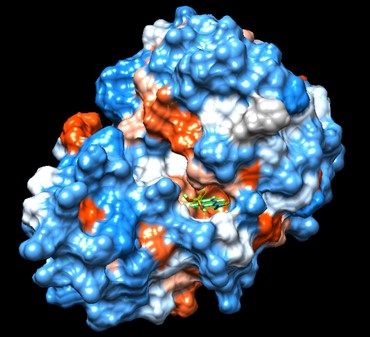
| - | == Structural highlights of 4DIU == | + | == Structural highlights of 4DIU ==<ref>PMID:21638687</ref> |
| - | + | ||
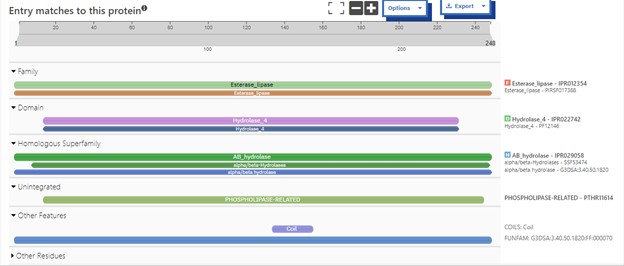
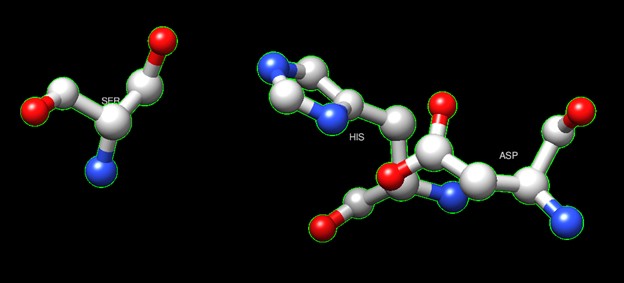
One of the distinctive structural characteristics of this protein is the alpha/beta-hydrolase (ABH) fold. Evidence for this fold consists of its structure (an open β-sheet surrounded by α-helices) and the presence of what is known as the "catalytic triad" (consisting of a nucleophile, an acid, and histidine)<ref>Holmquist, M. Alpha Beta-Hydrolase Fold Enzymes Structures, Functions and Mechanisms. Current Protein and Peptide Science 2000, 1 (2), 209–235. https://doi.org/10.2174/1389203003381405.</ref>. The presence of the active site residues His A 222, Asp A 192, and Ser A 93, as determined by SPRITE, Chimera, etc., confirms the presence of this catalytic triad. | One of the distinctive structural characteristics of this protein is the alpha/beta-hydrolase (ABH) fold. Evidence for this fold consists of its structure (an open β-sheet surrounded by α-helices) and the presence of what is known as the "catalytic triad" (consisting of a nucleophile, an acid, and histidine)<ref>Holmquist, M. Alpha Beta-Hydrolase Fold Enzymes Structures, Functions and Mechanisms. Current Protein and Peptide Science 2000, 1 (2), 209–235. https://doi.org/10.2174/1389203003381405.</ref>. The presence of the active site residues His A 222, Asp A 192, and Ser A 93, as determined by SPRITE, Chimera, etc., confirms the presence of this catalytic triad. | ||
Revision as of 16:32, 29 April 2024
| |||||||||||
References
- ↑ Fukami, T.; Yokoi, T. The Emerging Role of Human Esterases. Drug Metabolism and Pharmacokinetics 2012, 27 (5), 466–477. https://doi.org/10.2133/dmpk.dmpk-12-rv-042.
- ↑ Tokudome, Y.; Katayanagi, M.; Hashimoto, F. Esterase Activity and Intracellular Localization in Reconstructed Human Epidermal Cultured Skin Models. Annals of Dermatology 2015, 27 (3), 269. https://doi.org/10.5021/ad.2015.27.3.269.
- ↑ Williams, F. M. Clinical Significance of Esterases in Man. Clinical pharmacokinetics 1985, 10 (5), 392–403. https://doi.org/10.2165/00003088-198510050-00002.
- ↑ Zhang, S.; Sun, W.; Xu, L.; Zheng, X.; Chu, X.; Tian, J.; Wu, N.; Fan, Y. Identification of the Para-Nitrophenol Catabolic Pathway, and Characterization of Three Enzymes Involved in the Hydroquinone Pathway, in Pseudomonas Sp. 1-7. BMC Microbiology 2012, 12 (1). https://doi.org/10.1186/1471-2180-12-27.
- ↑ Vázquez-Mayorga, E.; Díaz-Sánchez, Á.; Dagda, R.; Domínguez-Solís, C.; Dagda, R.; Coronado-Ramírez, C.; Martínez-Martínez, A. Novel Redox-Dependent Esterase Activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson’s Disease. International Journal of Molecular Sciences 2016, 17 (8), 1346. https://doi.org/10.3390/ijms17081346.
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Holmquist, M. Alpha Beta-Hydrolase Fold Enzymes Structures, Functions and Mechanisms. Current Protein and Peptide Science 2000, 1 (2), 209–235. https://doi.org/10.2174/1389203003381405.