User:Melissa Siolin Martins/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
Ohr is unique when compared to other thiol based peroxidases, because it is very efficient in reducing OHPs but not so much when it comes to H2O2, while other peroxidases are broadspectrum, reducing OHP, H2O2 and other substrates such as fatty acid hydroperoxides and peroxynitrite with similar efficiencies. Therefore, Ohrs are specialized in OHPs, characteristic associated with the presence of a hydrophobic collar (HC) in their structure surrounding the active site, that interact more with hydrophobic substrates <ref name= "Meireles"/>. | Ohr is unique when compared to other thiol based peroxidases, because it is very efficient in reducing OHPs but not so much when it comes to H2O2, while other peroxidases are broadspectrum, reducing OHP, H2O2 and other substrates such as fatty acid hydroperoxides and peroxynitrite with similar efficiencies. Therefore, Ohrs are specialized in OHPs, characteristic associated with the presence of a hydrophobic collar (HC) in their structure surrounding the active site, that interact more with hydrophobic substrates <ref name= "Meireles"/>. | ||
| - | Ohr was described as a second system involved in bacterial resistance to OHPs by the discovery of the ohr gene from ''Xanthomonas campestris pv. phaseoli'' <ref name= "Meireles"/>. Furthermore, the | + | Ohr was described as a second system involved in bacterial resistance to OHPs by the discovery of the ohr gene from ''Xanthomonas campestris pv. phaseoli'' <ref name= "Meireles"/>. Furthermore, the Δohr mutant of ''X. campestris pv. phaseoli'' displayed a unique phenotype: increased and specific sensitivity to artificial OHPs but not to either H2O2 or superoxide generators. Ohr homologues from other bacteria such as ''Pseudomonas aeruginosa'' and ''Bacillus subtilis'' were also described as displaying a similar expression profile and role on OHP resistance <ref name= "Meireles"/>. |
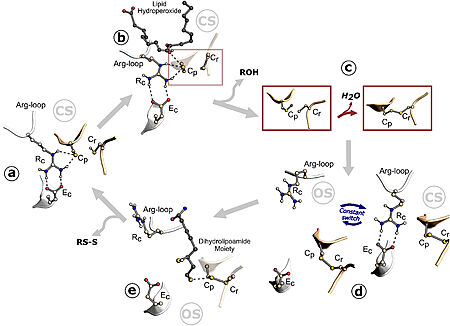
This enzime is part of the Ohr/OsmC protein family. This group is characterized for having a Cys-based thiol-dependent peroxidase activity. Besides not having much sequence similarities, their protein structure is highly conserved, presenting two Cys residues (Cp and Cr) in their active site. The other catalytic residues (Rc and Ec) are conserved among all of the Ohr/OsmC protein family members, but were not found in Ohr-like enzymes <ref name= "Alegria"/><ref name= "Meireles"/><ref name="Domingos">PMID:28391181</ref>. | This enzime is part of the Ohr/OsmC protein family. This group is characterized for having a Cys-based thiol-dependent peroxidase activity. Besides not having much sequence similarities, their protein structure is highly conserved, presenting two Cys residues (Cp and Cr) in their active site. The other catalytic residues (Rc and Ec) are conserved among all of the Ohr/OsmC protein family members, but were not found in Ohr-like enzymes <ref name= "Alegria"/><ref name= "Meireles"/><ref name="Domingos">PMID:28391181</ref>. | ||
| Line 27: | Line 27: | ||
The Ohr protein possesses an alpha/beta fold structure not found in peroxiredoxins or glutathione peroxidases <ref name="Alegria"/>. Ohr homologs and OsmC have motifs composed of well-conserved cysteine residues. One of these residues is part of a <scene name='10/1049474/Vcp_motif/1'>VCP motif</scene>, also found in peroxiredoxins, showing that Ohr can also participate in the peroxide decomposition process <ref name="cussiol">PMID:12540833</ref>. | The Ohr protein possesses an alpha/beta fold structure not found in peroxiredoxins or glutathione peroxidases <ref name="Alegria"/>. Ohr homologs and OsmC have motifs composed of well-conserved cysteine residues. One of these residues is part of a <scene name='10/1049474/Vcp_motif/1'>VCP motif</scene>, also found in peroxiredoxins, showing that Ohr can also participate in the peroxide decomposition process <ref name="cussiol">PMID:12540833</ref>. | ||
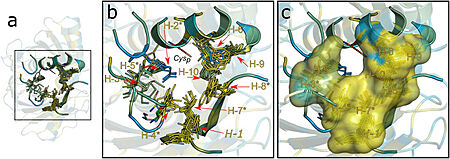
| - | Ohrs have a barrel-like structure composed of folded homodimers, in which two six-stranded β-sheets wrap around two central α-helices. In this conformation, there are two active sites at opposite sides of the dimer, being two Cp residues placed in the middle of one of the central α-helices <ref name= "Meireles"/>. The Ohr proteins catalytic mechanism of hydroperoxide reduction is setted on a pair of redox-active cysteines, named peroxidatic (Cp) and resolving (Cr) cysteines, resembling that of the atypical 2-Cys Prxs. Ohr acts like a dimer, and the cysteine residues are placed in opposing sides of each monomer as part of two, identical active sites <ref name= "Domingos"/>. | + | Ohrs have a barrel-like structure composed of folded homodimers, in which two six-stranded β-sheets wrap around two central <scene name='10/1049474/Alpha_helices/1'>α-helices</scene>. In this conformation, there are two active sites at opposite sides of the dimer, being two Cp residues placed in the middle of one of the central α-helices <ref name= "Meireles"/>. The Ohr proteins catalytic mechanism of hydroperoxide reduction is setted on a pair of redox-active cysteines, named peroxidatic (Cp) and resolving (Cr) cysteines, resembling that of the atypical 2-Cys Prxs. Ohr acts like a dimer, and the cysteine residues are placed in opposing sides of each monomer as part of two, identical active sites <ref name= "Domingos"/>. |
This structural analysis and amino acid sequence alignments showed the placement of a hydrophobic collar (HC) surrounding the active site of the protein. This part of the structure makes sense considering the fact that hydrophobicity is a common property among most Ohr substrates, such as dihydrolipoamide and lipoyl groups, both fatty acid hydroperoxides that are hydrophobic molecules <ref name= "Meireles"/>. | This structural analysis and amino acid sequence alignments showed the placement of a hydrophobic collar (HC) surrounding the active site of the protein. This part of the structure makes sense considering the fact that hydrophobicity is a common property among most Ohr substrates, such as dihydrolipoamide and lipoyl groups, both fatty acid hydroperoxides that are hydrophobic molecules <ref name= "Meireles"/>. | ||
| Line 36: | Line 36: | ||
Thus, in a simplified manner, due to the interactions between the amino acids of the active site, Ohr reduces its substrates to their corresponding products. To reduce more hydroperoxide molecules, it is necessary for the disulfide of this enzyme to be continuously reduced by other compounds. There is evidence that this role is performed by lipoylated proteins <ref name= "Alegria"/><ref name= "cussiol"/> | Thus, in a simplified manner, due to the interactions between the amino acids of the active site, Ohr reduces its substrates to their corresponding products. To reduce more hydroperoxide molecules, it is necessary for the disulfide of this enzyme to be continuously reduced by other compounds. There is evidence that this role is performed by lipoylated proteins <ref name= "Alegria"/><ref name= "cussiol"/> | ||
| + | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 18:29, 2 June 2024
Ohr protein
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Cussiol JR, Alegria TG, Szweda LI, Netto LE. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J Biol Chem. 2010 Jul 16;285(29):21943-50. doi: 10.1074/jbc.M110.117283. Epub, 2010 May 12. PMID:20463026 doi:http://dx.doi.org/10.1074/jbc.M110.117283
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Meireles DA, da Silva Neto JF, Domingos RM, Alegria TGP, Santos LCM, Netto LES. Ohr catalysis, phylogeny, regulation, and physiological roles. Free Radic Biol Med. 2022 May 20;185:6-24. PMID:35452809 doi:10.1016/j.freeradbiomed.2022.04.001
- ↑ Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1998 May;180(10):2636-43. PMID:9573147
- ↑ Lobo V, Patil A, Phatak A, Chandra N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn Rev. 2010 Jul;4(8):118-26. PMID:22228951 doi:10.4103/0973-7847.70902
- ↑ 5.0 5.1 Chen SJ, Shu HY, Lin GH. Regulation of tert-Butyl Hydroperoxide Resistance by Chromosomal OhrR in A. baumannii ATCC 19606. Microorganisms. 2021 Mar 18;9(3):629. PMID:33803549 doi:10.3390/microorganisms9030629
- ↑ 6.0 6.1 6.2 Meireles DA, Domingos RM, Gaiarsa JW, Ragnoni EG, Bannitz-Fernandes R, da Silva Neto JF, de Souza RF, Netto LES. Functional and evolutionary characterization of Ohr proteins in eukaryotes reveals many active homologs among pathogenic fungi. Redox Biol. 2017 Aug;12:600-609. PMID:28391181 doi:10.1016/j.redox.2017.03.026
- ↑ Alegria TG, Meireles DA, Cussiol JR, Hugo M, Trujillo M, de Oliveira MA, Miyamoto S, Queiroz RF, Valadares NF, Garratt RC, Radi R, Di Mascio P, Augusto O, Netto LE. Ohr plays a central role in bacterial responses against fatty acid hydroperoxides and peroxynitrite. Proc Natl Acad Sci U S A. 2017 Jan 10;114(2):E132-E141. PMID:28028230 doi:10.1073/pnas.1619659114
- ↑ 8.0 8.1 Cussiol JR, Alves SV, de Oliveira MA, Netto LE. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J Biol Chem. 2003 Mar 28;278(13):11570-8. doi: 10.1074/jbc.M300252200. Epub 2003 , Jan 22. PMID:12540833 doi:http://dx.doi.org/10.1074/jbc.M300252200
- ↑ Renato M. Domingos, Raphael D. Teixeira, Ari Zeida, William A. Agudelo, Thiago G. P. Alegria, José F. da Silva Neto, Plínio S. Vieira, Mario T. Murakami, Chuck S. Farah, Dario A. Estrin, and Luis E. S. Netto ACS Catalysis 2020 10 (12), 6587-6602. [DOI: 10.1021/acscatal.0c01257]