Arabidospsis thaliana PrRs (pinoresinol/lariciresinol reductases)
From Proteopedia
| Line 24: | Line 24: | ||
At the N-terminal end, each subunit of PrR contains a NADPH binding domain (NBD) formed by seven β-strands surrounded by six α-helices. The C-terminal end comprises the substrate binding domains (SBD), which is formed by two β-strands and five α-helices. In NDB, α-helices are larger than in SBD. Between both domains is a large groove containing a positively charged region that associates with NBD and a hydrophobic region that associates with SBD. Overall, the catalysis process consists of the binding of free NADPH to NBD, followed by binding of pinoresinol to SBD. The substrate is reduced and lariciresinol is released (Xiao et al., 2021). | At the N-terminal end, each subunit of PrR contains a NADPH binding domain (NBD) formed by seven β-strands surrounded by six α-helices. The C-terminal end comprises the substrate binding domains (SBD), which is formed by two β-strands and five α-helices. In NDB, α-helices are larger than in SBD. Between both domains is a large groove containing a positively charged region that associates with NBD and a hydrophobic region that associates with SBD. Overall, the catalysis process consists of the binding of free NADPH to NBD, followed by binding of pinoresinol to SBD. The substrate is reduced and lariciresinol is released (Xiao et al., 2021). | ||
| - | A flexible loop structure is formed by β-strand 4 (β4) and acts as a switch to control the binding of NADPH and release of NADP+ at NBD: without ligand β4 is disordered, but when PrR is binded with NADP+and pinoresinol it forms a well defined loop. Both monomer loops form a twisted “8” shape that covers NBD and SBD. Furthermore, NADPH binding causes the β2 loop to move slightly towards the coenzyme. NADP+ interacts with PrR grooves through hydrogen bonds and hydrophobic interactions with the NADPH-binding domain (<scene name='10/1050287/Gxxgxxg/ | + | A flexible loop structure is formed by β-strand 4 (β4) and acts as a switch to control the binding of NADPH and release of NADP+ at NBD: without ligand β4 is disordered, but when PrR is binded with NADP+and pinoresinol it forms a well defined loop. Both monomer loops form a twisted “8” shape that covers NBD and SBD. Furthermore, NADPH binding causes the β2 loop to move slightly towards the coenzyme. NADP+ interacts with PrR grooves through hydrogen bonds and hydrophobic interactions with the NADPH-binding domain (<scene name='10/1050287/Gxxgxxg/2'>GXXGXXG</scene>) and neighbor amino-acid residues (Xiao et al. 2021), being Phe166 responsible for the its stabilization (Markulin et al., 2019). |
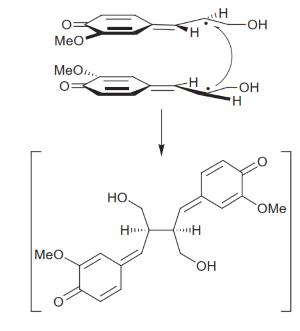
Pinoresinol binds to the hydrophobic portion of PrR grooves, being stabilized by β2 and α-helice 10 (α10). The binding of pinoresinol to one monomer induces a conformational change in the neighboring monomer that helps stabilize the substrate, resulting in a tighter bond. The substrate’s ends form hydrogen bonds with PrP Met125 and Gly178 residues, hence pinoresinol is bound to the enzyme as a straight chain. With this conformation, both molecules' hydrophobic regions are aligned (Xiao et al., 2021). Moreover, pinoresinol binds in such a way that its furan ring is facing the nicotinamide ring of the bound NADPH (Markulin et al., 2019), which allows the transference of an hydrogen atom to the substrate, reducing it to make one molecule of lariciresinol (Xiao et al., 2021). | Pinoresinol binds to the hydrophobic portion of PrR grooves, being stabilized by β2 and α-helice 10 (α10). The binding of pinoresinol to one monomer induces a conformational change in the neighboring monomer that helps stabilize the substrate, resulting in a tighter bond. The substrate’s ends form hydrogen bonds with PrP Met125 and Gly178 residues, hence pinoresinol is bound to the enzyme as a straight chain. With this conformation, both molecules' hydrophobic regions are aligned (Xiao et al., 2021). Moreover, pinoresinol binds in such a way that its furan ring is facing the nicotinamide ring of the bound NADPH (Markulin et al., 2019), which allows the transference of an hydrogen atom to the substrate, reducing it to make one molecule of lariciresinol (Xiao et al., 2021). | ||
Revision as of 20:04, 2 June 2024
Página sobre a PrR1 e PrR2 de Arabidopsis feita por alunos da Biologia da USP São Paulo.
A. thaliana PrRs
| |||||||||||
4. AtPrR expression
Understanding the pattern of expression of the PLR family is key in the study of lignans, considering that the diversity of these secondary metabolites is due to the action of these enzymes, but also to their expression pattern (Xiao et al., 2021; Markulin et al., 2019), which dictates where in the plants and when a certain lignan will be produced.
The pattern of expression of the PLRs varies from plant to plant and it can be altered by different stresses and hormones (Markunil et al., 2019). In the case of Arabidopsis thaliana, both PrR1 and PrR2 are expressed in root, but only PrR1 is also expressed in stem. Zhao and collaborators (2014) have shown by co-expression analysis that PrR1 might be related to the deposition of secondary cell wall, as it clustered with genes related to: lignification, hemicellulose biosynthesis, cellulose synthesis, etc. This possibility was reinforced by transactivation assays in which the trans-activators factors MYB46 and SND1, both related to the deposition of secondary cell wall, interacted with the PrR1 promoter. On other hand, PrR2 clustered with a different set of genes, been close to genes not related to secondary cell wall deposition, as well to genes related to lignification and to the formation of the casparian strip.
What might look contradictory is the fact that PrR1 and PrR2 are coexpressed with lignin related genes and that pinoresinol, the first substrate of the PLR enzymes, is synthetized by the dimerization of two coniferyl alcohols, onde of the monomers that constitute the lignin polymer. We could imagine that the production of lignans would compete for substrates with the biosynthesis of lignin, and therefore the coexpression mentioned would be questionable. Zhao and collaborators (2014) have shown that mutants of A. thaliana without the function of the PrR1 enzyme show lower content of lignin, meaning that, somehow the production of lignans is important for lignification and related to lignin polymerization.
5. References
- ↑ Xiao Y, Shao K, Zhou J, Wang L, Ma X, Wu D, Yang Y, Chen J, Feng J, Qiu S, Lv Z, Zhang L, Zhang P, Chen W. Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases. Nat Commun. 2021 May 14;12(1):2828. PMID:33990581 doi:10.1038/s41467-021-23095-y
- ↑ Markulin L, Corbin C, Renouard S, Drouet S, Gutierrez L, Mateljak I, Auguin D, Hano C, Fuss E, Lainé E. Pinoresinol-lariciresinol reductases, key to the lignan synthesis in plants. Planta. 2019 Jun;249(6):1695-1714. PMID:30895445 doi:10.1007/s00425-019-03137-y
MARKULIN, L. et al. Pinoresinol–lariciresinol reductases, key to the lignan synthesis in plants. Planta, v. 249, n. 6, p. 1695–1714, 20 mar. 2019. PMID: 30895445. DOI: https://doi.org/10.1007/s00425-019-03137-y.
TOMOYUKI NAKATSUBO et al. Characterization of Arabidopsis thaliana Pinoresinol Reductase, a New Type of Enzyme Involved in Lignan Biosynthesis. Journal of Biological Chemistry, v. 283, n. 23, p. 15550–15557, 1 jun. 2008. PMID: 18347017. DOI: https://doi.org/10.1074%2Fjbc.M801131200.
WANG, L. et al. Review of lignans from 2019 to 2021: Newly reported compounds, diverse activities, structure-activity relationships and clinical applications. Phytochemistry, v. 202, p. 113326–113326, 1 out. 2022.
WARD, R. Lignans, neolignans and related compounds. v. 16, n. 1, p. 75–96, 1 jan. 1999. doi: https://doi.org/10.1039/A705992B
XIAO, Y. et al. Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases. Nature communications, v. 12, n. 1, 14 maio 2021. PMID: 33990581. DOI: https://doi.org/10.1038/s41467-021-23095-y.
ZHAO, Q. et al. Pinoresinol reductase 1 impacts lignin distribution during secondary cell wall biosynthesis in Arabidopsis. Phytochemistry, v. 112, p. 170–178, 1 abr. 2015. PMID: 25107662. DOI: https://doi.org/10.1016/j.phytochem.2014.07.008.
Proteopedia Page Contributors and Editors (what is this?)
Gabriel Fontanella Pileggi, Maisa Ganz Sanchez Sennes, Melissa Siolin Martins, Michal Harel, Angel Herraez