Journal:Acta Cryst D:S2059798324005229
From Proteopedia
(Difference between revisions)

| Line 6: | Line 6: | ||
Single particle cryo-EM has become a routine and indispensable technique in structural biology, where macromolecules are imaged with electrons as single particles and subsequently averaged to obtain a high-resolution 3D reconstruction. Single particle cryo-EM reconstruction utilizes the projection slice theorem to build a 3D map of a 3D object using 2D projections, generated when electrons transmitted through a thin specimen are recorded by a detector. The resolution and quality of the final map depend on many factors including heterogeneity in the specimen, adequate sampling of views and signal-to-noise of the images. | Single particle cryo-EM has become a routine and indispensable technique in structural biology, where macromolecules are imaged with electrons as single particles and subsequently averaged to obtain a high-resolution 3D reconstruction. Single particle cryo-EM reconstruction utilizes the projection slice theorem to build a 3D map of a 3D object using 2D projections, generated when electrons transmitted through a thin specimen are recorded by a detector. The resolution and quality of the final map depend on many factors including heterogeneity in the specimen, adequate sampling of views and signal-to-noise of the images. | ||
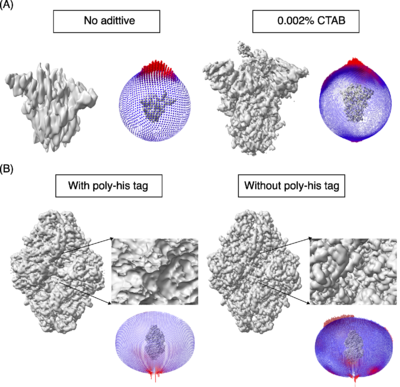
A key step in this process is to obtain a thin film of macromolecules (purified or as a mixture) in ice and the most routinely used method for obtaining such thin films was developed by Dubochet and his colleagues (Dubochet ''et al.,'' 1988<ref name="Dubochet1">PMID:3043536</ref>). Obtaining the optimal conditions for freezing of a given macromolecule requires testing several parameters. At the moment, it is one of the rate-limiting steps in obtaining high-resolution maps by cryo-EM. During the freezing process, the macromolecules can encounter and interact with the air-water interface, sometimes resulting in a tendency to adapt one or more preferential orientations along with denaturation and dissociation in the case of multi-subunit complexes (D’Imprima ''et al.,'' 2019<ref name="D’Imprima">PMID:30932812</ref>; Noble ''et al.,'' 2018<ref name="Noble">PMID: 30250056</ref>; Glaeser and Han, 2017<ref name="Glaeser">PMID: 28781996</ref>). The 3D maps reconstructed from preferentially oriented macromolecules exhibit stretching of densities in one direction (also called anisotropy, where with enough particles the resolution will be high but the maps are not easily interpretable). One such example is the spike protein shown in Figure 1A, which illustrates the effects of orientation bias on the final reconstruction. | A key step in this process is to obtain a thin film of macromolecules (purified or as a mixture) in ice and the most routinely used method for obtaining such thin films was developed by Dubochet and his colleagues (Dubochet ''et al.,'' 1988<ref name="Dubochet1">PMID:3043536</ref>). Obtaining the optimal conditions for freezing of a given macromolecule requires testing several parameters. At the moment, it is one of the rate-limiting steps in obtaining high-resolution maps by cryo-EM. During the freezing process, the macromolecules can encounter and interact with the air-water interface, sometimes resulting in a tendency to adapt one or more preferential orientations along with denaturation and dissociation in the case of multi-subunit complexes (D’Imprima ''et al.,'' 2019<ref name="D’Imprima">PMID:30932812</ref>; Noble ''et al.,'' 2018<ref name="Noble">PMID: 30250056</ref>; Glaeser and Han, 2017<ref name="Glaeser">PMID: 28781996</ref>). The 3D maps reconstructed from preferentially oriented macromolecules exhibit stretching of densities in one direction (also called anisotropy, where with enough particles the resolution will be high but the maps are not easily interpretable). One such example is the spike protein shown in Figure 1A, which illustrates the effects of orientation bias on the final reconstruction. | ||
| - | [[Image:Figure1RR5238.png|left|thumb|400px|'''Figure 1''']] | + | [[Image:Figure1RR5238.png|left|thumb|400px|'''Figure 1:''' The models used as reference are PDBs [[8h3d]] and [[6cvm]] for the spike protein and b-galactosidase respectively. A) Spike protein, SARS-COV-2. No additive. <scene name='10/1050322/Spike_with_ctab/1'>Spike with CTAB</scene>.]] |
{{Clear}} | {{Clear}} | ||
Revision as of 14:27, 10 June 2024
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.