Trypsin
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
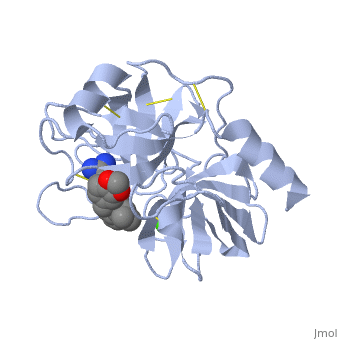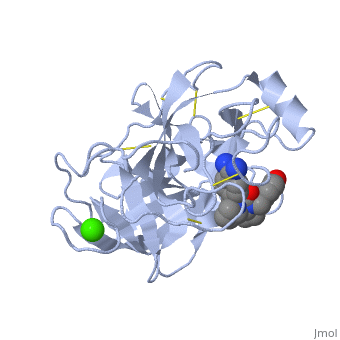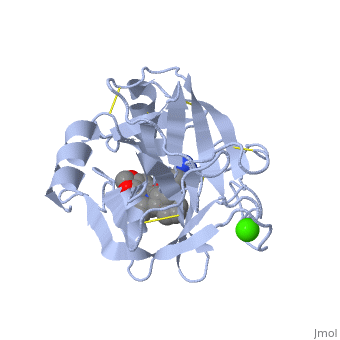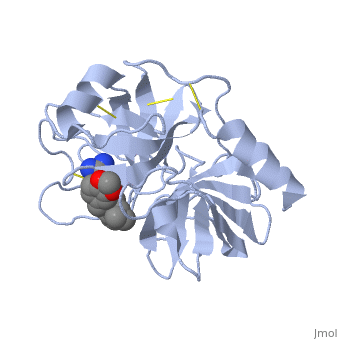
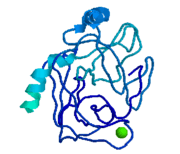
The binding of trypsin to UB-THR 10 somewhat emulates the binding to its specific peptide substrates. The preference for lysine or arginine in trypsin catalysis is due to the composition of the trypsin <scene name='10/100160/Cv/5'>specificity pocket</scene>. Here (green), Asp 189 and one of two significant glycine backbones, Gly 216, interact with the ligand as they would with Arg or Lys. | The binding of trypsin to UB-THR 10 somewhat emulates the binding to its specific peptide substrates. The preference for lysine or arginine in trypsin catalysis is due to the composition of the trypsin <scene name='10/100160/Cv/5'>specificity pocket</scene>. Here (green), Asp 189 and one of two significant glycine backbones, Gly 216, interact with the ligand as they would with Arg or Lys. | ||
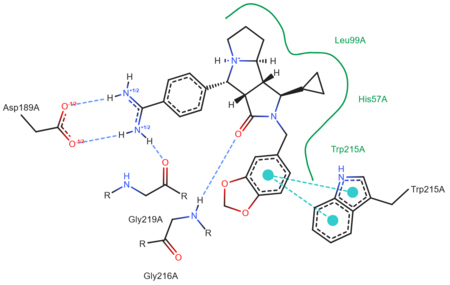
| - | [[Image: | + | [[Image:Ligand_1y3v_ProteinsPlus.png|thumb|left|upright=2.5|A two-dimensional representation of trypsin binding Ligand UB-THR 10]] |
{{Clear}} | {{Clear}} | ||
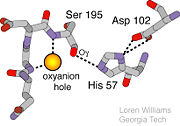
The <scene name='10/100160/Cv/6'>catalytic triad</scene>; Asp 102, His 57, and Ser 195, shown here in yellow, is positioned near the substrate. The catalytically active histidine and serine side chains are even near an amide bond in UB-THR 10, just like the amide bond broken in peptide hydrolysis. According to FirstGlance in Jmol, there is no bonding of these groups with the ligand, apart from minor van der Waal's interactions with Hist 57. If Ligand UB-Thr 10 were a transition state analog, some covalent connection would exist in addition to hydrogen bonds. UB-THR 10 simulates the substrate, but does not hydrolyze at either of its two amide bonds, likely due to the local cyclic groups atypical of peptide backbones. | The <scene name='10/100160/Cv/6'>catalytic triad</scene>; Asp 102, His 57, and Ser 195, shown here in yellow, is positioned near the substrate. The catalytically active histidine and serine side chains are even near an amide bond in UB-THR 10, just like the amide bond broken in peptide hydrolysis. According to FirstGlance in Jmol, there is no bonding of these groups with the ligand, apart from minor van der Waal's interactions with Hist 57. If Ligand UB-Thr 10 were a transition state analog, some covalent connection would exist in addition to hydrogen bonds. UB-THR 10 simulates the substrate, but does not hydrolyze at either of its two amide bonds, likely due to the local cyclic groups atypical of peptide backbones. | ||
Revision as of 20:54, 25 January 2025
| |||||||||||
References
- ↑ Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html>
- ↑ Trypsin. 30 October 2010 <http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzyme/trypsin.html>.
- ↑ Image From: http://chemistry.umeche.maine.edu/MAT500/Peptidase1.html
- ↑ Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html>
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.
- ↑ Image From:

- ↑ Williams, Loren. Georgia Tech. http://www2.chemistry.gatech.edu/~1W26/bcourse_information/6521/protein/serine_protease/triad_1/html.
- ↑ Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Eran Hodis, Leah Bowlin, David Canner, Karsten Theis, Glenn Jones, Ben Hallowell, Karl Oberholser, Jaime Prilusky