Sandbox Reserved 1849
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
==SARS-COV-2 Spike Protein== | ==SARS-COV-2 Spike Protein== | ||
| - | <div> | ||
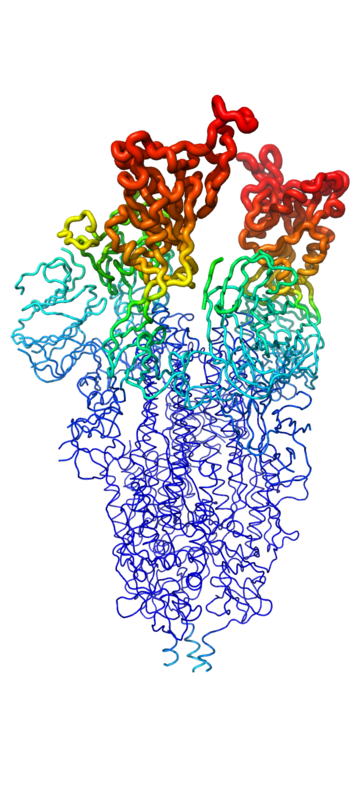
<p>The <scene name='10/1075251/Spike_protein_closed_spacefill/2'>spike protein</scene> of SARS-COV-2 is a symmetric trimer featuring 3 spike glycoprotein chains (UNIPROT: [https://www.uniprot.org/uniprotkb/P0DTC2/entry P0DTC2]). Each monomer of the spike is called a spike glycoprotein, and the total assembly contains 2 main parts: The <scene name='10/1075251/Spike_protein_1-up_yang_s1/1'>S1</scene> and <scene name='10/1075251/Spike_protein_1-up_yang_s2/1'>S2</scene> subunits<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref>. However, the native spike protein does not exist in this state prior to infection. The protein is actually inactive initially, but is later activated by proteases cleaving the inactive S protein into its two active subunits<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref>. The S1 subunit contains the <scene name='10/1075251/Spike_protein_2-up_yang_s1_dom/1'>N-Terminal domain (NTD), C-Terminal Domain (CTD), and the Receptor Binding Domain (RBD)</scene>. The RBD is responsible for binding to the ACE2 receptor on the surface of the target cell, as well as neutralizing antibodies. The NTD, CTD, and their relevant interfaces actually play much larger roles in the binding of the spike protein to ACE2 than the RBD does due to their larger surface areas<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref>. The S2 subunit is responsible for viral fusion and entry. Once bound to ACE2, and after the different domains in S2 have anchored to the membrane as well as delivered the viral envelope, the S2 subunit then changes conformation from the pre-hairpin to postfusion-hairpin conformation<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref>. The S2 subunit contains a fusion peptide domain (FP), heptapeptide repeat sequences 1 and 2 (HR1 & HR2), TM domain, and cytoplasmic fusion domain (CT). Full information about the location and structures of these domains within the S2 subunit can be found in references 1 and 3<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref><ref name="Zhang">PMID:34534731</ref>. For the purpose of this article about the minibinders, attention will be directed to the S1 subunit and its binding properties with ACE2. | <p>The <scene name='10/1075251/Spike_protein_closed_spacefill/2'>spike protein</scene> of SARS-COV-2 is a symmetric trimer featuring 3 spike glycoprotein chains (UNIPROT: [https://www.uniprot.org/uniprotkb/P0DTC2/entry P0DTC2]). Each monomer of the spike is called a spike glycoprotein, and the total assembly contains 2 main parts: The <scene name='10/1075251/Spike_protein_1-up_yang_s1/1'>S1</scene> and <scene name='10/1075251/Spike_protein_1-up_yang_s2/1'>S2</scene> subunits<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref>. However, the native spike protein does not exist in this state prior to infection. The protein is actually inactive initially, but is later activated by proteases cleaving the inactive S protein into its two active subunits<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref>. The S1 subunit contains the <scene name='10/1075251/Spike_protein_2-up_yang_s1_dom/1'>N-Terminal domain (NTD), C-Terminal Domain (CTD), and the Receptor Binding Domain (RBD)</scene>. The RBD is responsible for binding to the ACE2 receptor on the surface of the target cell, as well as neutralizing antibodies. The NTD, CTD, and their relevant interfaces actually play much larger roles in the binding of the spike protein to ACE2 than the RBD does due to their larger surface areas<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref>. The S2 subunit is responsible for viral fusion and entry. Once bound to ACE2, and after the different domains in S2 have anchored to the membrane as well as delivered the viral envelope, the S2 subunit then changes conformation from the pre-hairpin to postfusion-hairpin conformation<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref>. The S2 subunit contains a fusion peptide domain (FP), heptapeptide repeat sequences 1 and 2 (HR1 & HR2), TM domain, and cytoplasmic fusion domain (CT). Full information about the location and structures of these domains within the S2 subunit can be found in references 1 and 3<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref><ref name="Zhang">PMID:34534731</ref>. For the purpose of this article about the minibinders, attention will be directed to the S1 subunit and its binding properties with ACE2. | ||
| Line 13: | Line 12: | ||
Throughout the entire process, the spike protein has 3 main conformations. An <scene name='10/1075251/Spike_protein_closed_spacefill/3'>inactive, "closed"</scene> conformation; an active, "open" conformation; and a <scene name='10/1075251/Spike_protein_postfusion/1'>post-fusion hairpin</scene> conformation mentioned previously<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref><ref name="Yuan">PMID:28393837</ref><ref name="Zhang">PMID:34534731</ref>. In the closed conformation, the RBDs of each monomer are tucked inwards, preventing interaction. In the open conformation, however, 1 or more of these RBDs can be in the "up" conformation, meaning they are exposed and able to interact within the extracellular space. Mainly, there exits a "<scene name='10/1075251/Spike_protein_1-up_yang/1'>1-up</scene>" and "<scene name='10/1075251/Spike_protein_2-up_yang/2'>2-up</scene>" conformation in this phase<ref name="Yuan">PMID:28393837</ref><ref name="Zhang">PMID:34534731</ref>. Depicted in Figure 1, the RBDs of the spike protein have the highest mobility, which further support the many conformational changes in which they are involved. Most of the depictions of the minibinders bound to the spike protein show the spike protein in the 2-up conformation. | Throughout the entire process, the spike protein has 3 main conformations. An <scene name='10/1075251/Spike_protein_closed_spacefill/3'>inactive, "closed"</scene> conformation; an active, "open" conformation; and a <scene name='10/1075251/Spike_protein_postfusion/1'>post-fusion hairpin</scene> conformation mentioned previously<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref><ref name="Yuan">PMID:28393837</ref><ref name="Zhang">PMID:34534731</ref>. In the closed conformation, the RBDs of each monomer are tucked inwards, preventing interaction. In the open conformation, however, 1 or more of these RBDs can be in the "up" conformation, meaning they are exposed and able to interact within the extracellular space. Mainly, there exits a "<scene name='10/1075251/Spike_protein_1-up_yang/1'>1-up</scene>" and "<scene name='10/1075251/Spike_protein_2-up_yang/2'>2-up</scene>" conformation in this phase<ref name="Yuan">PMID:28393837</ref><ref name="Zhang">PMID:34534731</ref>. Depicted in Figure 1, the RBDs of the spike protein have the highest mobility, which further support the many conformational changes in which they are involved. Most of the depictions of the minibinders bound to the spike protein show the spike protein in the 2-up conformation. | ||
| - | </p | + | </p> |
==ACE2== | ==ACE2== | ||
ACE2 is a carboxypeptidase present on cell surfaces that is responsible for the degradation of angiotensin II. It is a critical enzyme in the suppression of the renin-angiotensin system. This improves both cardiovascular and renal systems, as well as abates acute respiratory distress syndrome (ARDS). It does the 2 former via the RAS System's role in the regulation of blood pressure, renal function, water homeostasis, electrolyte balance, and/or inflammation<ref name="Kuba ACE2-SARS Receptor Model">DOI:10.3389/fimmu.2021.732690</ref>. The critical role that this enzyme plays in the regulation of this system is what results in the adverse symptomology observed in victims of the SARS-COV-2 virus. The ACE2 receptor is considered the only essential receptor in the SARS-COV-2 viral mechanism, and thus the collateral debilitation of ACE2 results in the adverse respiratory effects including ARDS, pulmonary edema, destruction of alveolar structures, and others<ref name="Kuba ACE2-SARS Receptor Model">DOI:10.3389/fimmu.2021.732690</ref>. This relationship was further proven when ACE-2 deficient mice had developed these effects at higher rates compared to the wild type<ref name="Kuba Lung Injury">PMID:16007097</ref>. | ACE2 is a carboxypeptidase present on cell surfaces that is responsible for the degradation of angiotensin II. It is a critical enzyme in the suppression of the renin-angiotensin system. This improves both cardiovascular and renal systems, as well as abates acute respiratory distress syndrome (ARDS). It does the 2 former via the RAS System's role in the regulation of blood pressure, renal function, water homeostasis, electrolyte balance, and/or inflammation<ref name="Kuba ACE2-SARS Receptor Model">DOI:10.3389/fimmu.2021.732690</ref>. The critical role that this enzyme plays in the regulation of this system is what results in the adverse symptomology observed in victims of the SARS-COV-2 virus. The ACE2 receptor is considered the only essential receptor in the SARS-COV-2 viral mechanism, and thus the collateral debilitation of ACE2 results in the adverse respiratory effects including ARDS, pulmonary edema, destruction of alveolar structures, and others<ref name="Kuba ACE2-SARS Receptor Model">DOI:10.3389/fimmu.2021.732690</ref>. This relationship was further proven when ACE-2 deficient mice had developed these effects at higher rates compared to the wild type<ref name="Kuba Lung Injury">PMID:16007097</ref>. | ||
| + | |||
| + | As mentioned previously, all of the S1 subunit domains play important roles in the binding to ACE2. The surface area of the NTD and CTD are particularly important, along with the direct interactions observed in the RBD. Whilst ACE2 is not the focus of this article, understanding its role in the infection pathway of COVID 19, as well as how it binds to the spike protein will assist in understanding the design and functional processes of the minibinders. | ||
==Minibinders== | ==Minibinders== | ||
===Structure=== | ===Structure=== | ||
Revision as of 19:36, 3 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
SARS-COV2 Minibinders
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020 Sep;41(9):1141-1149. doi: 10.1038/s41401-020-0485-4., Epub 2020 Aug 3. PMID:32747721 doi:http://dx.doi.org/10.1038/s41401-020-0485-4
- ↑ 4.0 4.1 4.2 4.3 Zhang J, Xiao T, Cai Y, Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol. 2021 Oct;50:173-182. PMID:34534731 doi:10.1016/j.coviro.2021.08.010
- ↑ 5.0 5.1 5.2 Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017 Apr 10;8:15092. doi: 10.1038/ncomms15092. PMID:28393837 doi:http://dx.doi.org/10.1038/ncomms15092
- ↑ 6.0 6.1 6.2 Kuba K, Yamaguchi T, Penninger JM. Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19. Front Immunol. 2021 Dec 22;12:732690. PMID:35003058 doi:10.3389/fimmu.2021.732690
- ↑ 7.0 7.1 Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005 Aug;11(8):875-9. PMID:16007097 doi:10.1038/nm1267