User:Emily Hwang/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 55: | Line 55: | ||
[[Image:PET_hydrolase_Mechanism.jpeg|400 px|left|thumb|Figure Legend]] <b>need higher quality image of this</b> | [[Image:PET_hydrolase_Mechanism.jpeg|400 px|left|thumb|Figure Legend]] <b>need higher quality image of this</b> | ||
==Mutations== | ==Mutations== | ||
| + | Researchers have been investigating various mutations of PET hydrolase to enhance its catalytic ability. One group of researchers, Tournier et. al., have made mutations in the PET hydrolase active site. They identified the key residues involved in the catalytic mechanism by using a model of the <scene name='10/1075190/Ligand/2'>PET substrate</scene> (2-HE(MHET)₃) onto the enzyme (PDB ID 4EB0). The site, mainly a hydrophobic pocket, contained 11 residues targeted for mutagenesis. From this, they identified that the majority of enzymes' specific activity went down; however, the mutation of the <scene name='10/1075190/F243_wt/5'>WT F243 residue</scene> to either isoleucine or tryptophan increased specific activity. | ||
| + | |||
| + | The mutation of the F243 position to a tryptophan (<scene name='10/1075190/F243w_mutant/6'>F243W</scene>) was selected for further analysis based on its enhanced catalytic activity in the depolymerization of Pf-PET. The W243 mutation was shown to improve the substrate's binding affinity and increase the enzyme’s activity compared to the wild-type enzyme. This was one of the few variants that exhibited higher activity, and it was further analyzed through differential scanning fluorimetry (DSF) to assess its stability.<ref name="Tournier et. al. 2020">PMID:32269349</ref> | ||
| + | |||
| + | Similar to the W243 mutation, the <scene name='10/1075190/F243i/4'>F234I</scene> mutation was identified as a variant with improved depolymerization activity of Pf-PET. The I243 mutation in LCC led to better substrate interaction than the wild-type enzyme. After generating all possible variants, this mutation was among the few that exhibited 75% or more of the wild-type specific activity. As with the W243 mutation, DSF was used to determine the melting temperature and thermal stability, supporting the increased activity observed with this mutation. | ||
| + | |||
| + | == Results == | ||
| + | '''Table 1.''' Specific activity and rate of wild-type, WCCG, and ICCG mutants. Initial rate was measured by the calculated rate of reaction by NaOH consumption. Specific activity was measured via pf-PET-depolymerization assay.<ref name="Tournier et. al. 2020">PMID:32269349</ref> | ||
| + | |||
| + | <table> | ||
| + | <tr> | ||
| + | <th style="width:0%"><th>Enzyme</th> | ||
| + | <th style="width:5%"><th>Initial rate (g<sub>hydrolyzed PET</sub>•L<sup>-1</sup>•h<sup>-1</sup>)</th> | ||
| + | <th style="width:5%"><th>Specific activity (mg<sub>TAeq</sub>h<sup>-1</sup>•mg<sub>enzyme</sub><sup>-1</sup>) ± SD </th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th style="width:0%"><td>Wild-Type</td> | ||
| + | <th style="width:10%"><td>25.7</td> | ||
| + | <th style="width:10%"><td>81.9 ± 5.6 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th style="width:0%"><td>WCCG</td> | ||
| + | <th style="width:10%"><td>30.3</td> | ||
| + | <th style="width:10%"><td>75.9 ± 5.9</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th style="width:0%"><td>ICCG</td> | ||
| + | <th style="width:10%"><td>31.0</td> | ||
| + | <th style="width:10%"><td>82.0 ± 3.9</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | Both the WCCG and ICCG mutants display slightly higher initial rates (30.3 and 31.0 g hydrolyzed PET•L⁻¹•h⁻¹, respectively) compared to the wild-type enzyme (25.7 g hydrolyzed PET•L⁻¹•h⁻¹). This suggests that the mutations introduced in WCCG and ICCG enhance the rate of PET breakdown. | ||
| + | |||
| + | The specific activity of the WCCG mutant (75.9 mg TAeq h⁻¹ mg⁻¹ enzyme) is slightly lower than that of wild-type PETase (81.9 mg TAeq h⁻¹ mg⁻¹ enzyme), whereas the ICCG mutant shows comparable specific activity to the wild-type (82.0 mg TAeq h⁻¹ mg⁻¹ enzyme). | ||
| + | |||
| + | == Mechanism == | ||
| + | |||
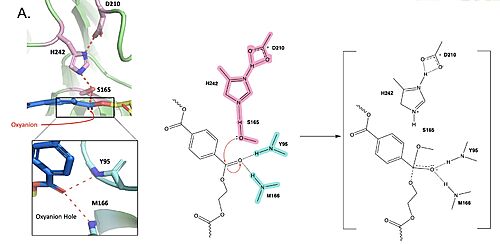
| + | Polyethylene terephthalate (PET) hydrolase (PETase) is a serine hydrolase that catalyzes the cleavage of ester bonds in PET polymers. | ||
| + | |||
| + | <center>[[Image:PartImechaism.jpg|500 px|center|thumb|Figure 1. Active site of PETase highlighting catalytic residues and the first step of the reaction mechanism. Residues in pink represent the catalytic triad (S165, H242, and D210). Light blue represents the oxyanion hole residues (Y95 and M166).]]</center> | ||
| + | In the first step of the reaction, the nucleophilic serine residue (S165) attacks the carbonyl carbon of the PET substrate, forming a tetrahedral intermediate. The negative charge is stabilized by the oxyanion hole. | ||
| + | |||
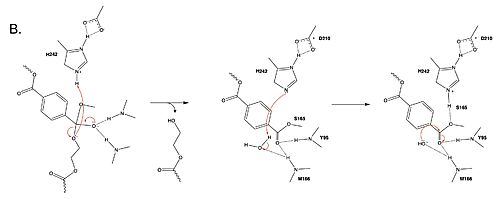
| + | <br> <center>[[Image:Part2mech.jpg|500 px|center|thumb|Figure 2. Second step of the PET hydrolysis reaction by PETase.]]</center> | ||
| + | The collapse of the tetrahedral intermediate reforms the double bond at the carbonyl, while the catalytic histidine (H242) protonates the leaving group monomer of the PET substrate. H242 then activates a water molecule as a nucleophile to attack the intermediate at S165. | ||
| + | |||
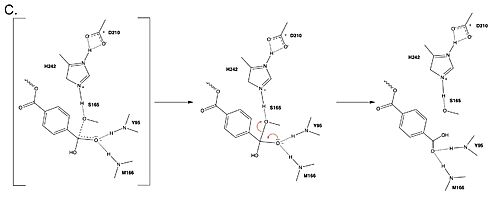
| + | <br> <center>[[Image:Part3Mech.jpg|500 px|center|thumb|Figure 3. Final step of PET hydrolysis by PETase.]]</center> | ||
| + | In the final step, the water-activated nucleophile attacks the carbonyl carbon of S165, generating a second tetrahedral intermediate. The collapse of this intermediate regenerates the carbonyl double bond and removes the catalytic serine residue. The oxyanion hole residues stabilize the negative charge of this transition state, allowing for product release of the PET monomer. | ||
| + | |||
Researchers have been investigating various mutations of PET hydrolase to enhance its catalytic ability. One group of researchers, Tournier et. al., have made mutations in the PET hydrolase active site. They identified the key residues involved in the catalytic mechanism by using a model of the <scene name='10/1075190/Ligand/2'>PET substrate</scene> onto the enzyme. The site, mainly a hydrophobic pocket, contained 11 residues targeted for mutagenesis. From this, they identified that the majority of enzymes' specific activity went down; however, the mutation of the F243 to either isoleucine or tryptophan increased specific activity. Four target mutations introduced into the PET Hydrolase by Tournier et. al. demonstrated improved catalytic efficiency and thermal stability compared to its wild-type structure. | Researchers have been investigating various mutations of PET hydrolase to enhance its catalytic ability. One group of researchers, Tournier et. al., have made mutations in the PET hydrolase active site. They identified the key residues involved in the catalytic mechanism by using a model of the <scene name='10/1075190/Ligand/2'>PET substrate</scene> onto the enzyme. The site, mainly a hydrophobic pocket, contained 11 residues targeted for mutagenesis. From this, they identified that the majority of enzymes' specific activity went down; however, the mutation of the F243 to either isoleucine or tryptophan increased specific activity. Four target mutations introduced into the PET Hydrolase by Tournier et. al. demonstrated improved catalytic efficiency and thermal stability compared to its wild-type structure. | ||
===F243I/W Mutations=== | ===F243I/W Mutations=== | ||
Revision as of 16:48, 12 April 2025
| |||||||||||
References
- ↑ 1.0 1.1 Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ 2.0 2.1 2.2 2.3 Shirke AN, White C, Englaender JA, Zwarycz A, Butterfoss GL, Linhardt RJ, Gross RA. Stabilizing Leaf and Branch Compost Cutinase (LCC) with Glycosylation: Mechanism and Effect on PET Hydrolysis. Biochemistry. 2018 Feb 20;57(7):1190-1200. PMID:29328676 doi:10.1021/acs.biochem.7b01189
- ↑ Imperiali B, O'Connor SE. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr Opin Chem Biol. 1999 Dec;3(6):643-9. PMID:10600722
Student Contributors
- Georgia Apple
- Emily Hwang
- Anjali Rabindran