We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Shea Bailey/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
===Active Site=== | ===Active Site=== | ||
| - | <scene name='10/1075219/ | + | In order to best target the RBD of the spike protein, the minibinders reveal a wide range of interactions to compete with <scene name='10/1075219/Ace2/1'>ACE2 binding</scene>. |
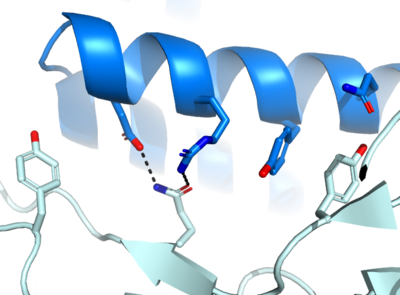
| - | <scene name='10/1075219/ | + | The first design method, Rosetta, created AHB2 based on the single interacting helix of ACE2. With <scene name='10/1075219/Ahb2/2'>AHB2 binding</scene>, we see two alpha helices mimicking ACE2. Hydrogen bonding interactions between N36, D11, K43, E41, and E30 of the minibinder interact with residues K417, R403, Y449, Q493, and N487, respectively, in the spike protein. |
| - | <scene name='10/1075219/ | + | De novo designed proteins, as discussed previously, focused on computational design to determine residues best able to interact with the spike protein. We will focus on LCB1 and LCB3. <scene name='10/1075219/Lcb1/4'>LCB1 binding</scene> reveals hydrogen bonding between D30 of the minibinder and both K417 and R403 of the spike protein, in addition to D17 and R14 of the minibinder interacting with Q493 of the spike protein. Similarly, <scene name='10/1075219/Lcb3/5'>LCB3 binding</scene> reveals hydrogen bonding between D11 of the minibinder to K417 and R403 of the spike protein. |
| - | + | Within all four binding sites, we see two conserved residues throughout: K417 and Q493. | |
</StructureSection> | </StructureSection> | ||
Revision as of 17:49, 13 April 2025
==SARS-CoV-2 Miniprotein Inhibitors, LCB1==
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park YJ, Strauch EM, Stewart L, Diamond MS, Veesler D, Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020 Oct 23;370(6515):426-431. PMID:32907861 doi:10.1126/science.abd9909
Student Contributors
- Shea Bailey