Introduction
Impact of Scaffold Rigidity
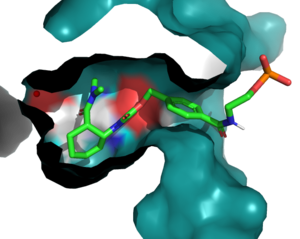
The Diels-Alderase protein aims to create optimal reacting conditions between the diene and dienophile in a Diels-Alder reaction. It accomplishes this by decreasing the energy gap between the dienophile’s lowest unoccupied molecular orbital (LUMO) and the diene’s highest occupied molecular orbital (HOMO) in the transition state.[1] The binding pocket of 4O5T is selective for two substrates, 4-carboxybenzyl trans-1,3-butadiene-1-carbamate (diene) and N,N- dimethylacrylamide (dienophile). The binding site contains an H-bond donor (Y134) which lowers the LUMO energy and stabilizes the negative charge on the dienophile and an H-bond acceptor (Q208) that increases the HOMO energy and stabilizes the positive charge on the diene. Both of these H-bonding interactions work to stabilize the transition state, while also orienting the substrates in optimal conformations for reacting.
General Structure
Active Site
In the active state, there are that aim to stabilize the transition state of the Diels-Alder reaction. The Y134 acts as a to the oxygen on the . Q208 acts as a to the nitrogen on the ligand. These interactions help reduce the energetic gap between orbitals allowing the reaction to proceed, outlined in HOMO/LUMO.
Helix Cap
In the evolution process, researchers added a 16-residue a-helix motif to the top of the binding site. The hydrophobic helix “functions as a lid to constrain the substrates in a productive orientation for reaction” (CITE), decreasing the Km of the enzyme and increasing the catalytic efficiency, as seen in the measured kinetics of the enzyme.
Mechanism
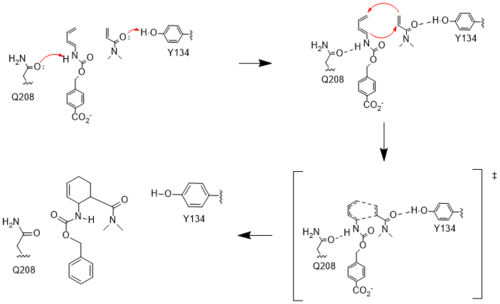
Figure #. Active site mechanism
HUMO and LUMO
The two active site residues, Y134 and Q208, use hydrogen bonding to close the energy gap between the HOMO diene and the LUMO dienophile. The goal of closing the energy gap is so that in the diels-alder reaction, the diene and dienophile can readily switch roles for the mechanism to progress and complete the formation of the product. Due to the conserved nature of this mechanism, the diels-alderase is stereoselective for the 3R, 4S endo product.
Hydrogen Binding
Rather than using acid/base catalysis like many enzymes, the diels-alderase utilized hydrogen bonding to alter the HOMO and LUMO energies of the diene and dienophile. Y134 donates a hydrogen bond to the dieneophile, increasing the electron density and lowering the LUMO. Q208 accepts a hydrogen bond from the diene, decreasing the electron density and lowering the HOMO.
Development and Evolution
DA_20_10
DA_20_10 provided key mutations in and around the active site that increased the hydrophobicity, provided structural stability, and increased interactions between the ligand and surrounding residues.
Q162R
- Residue 162 resides near the top of the binding entrance to the enzyme, and is within 3A in most models on the enzyme. It can act as a hydrogen bond donor to the terminal phosphate on the ligand when in proximity. To increase this interaction, the group chose a Q to , which decreased the length of the potential hydrogen bond to within 2.5A, increasing the strength of the interaction.
S284A
Residue 284 resides deep within the binding pocket of the enzyme. The group chose an to mutation to increase the hydrophobicity of the binding pocket and reduce reactivity, without also changing any steric characteristics in the region unintentionally near the catalytic residues.
A285N
- , as follows, is also buried within the binding pocket. The group introduced this mutation to increase steric hindrance with the catalytic tyrosine, reducing the number of rotamers the residue has to increase the reactivity of the enzyme by lowering the distance between Y134 and the ligand.
CE6
The DA_20_10 model of the Diels Alderase was further enhanced by players of the online game "Foldit." Building on preliminary early data, players were asked to optimize various helical structures that would surround and support the ligand. After over 100,000 designs were tested, the top-scoring CE6 model was finalized, containing as that favorably constrains ligand orientation. This "cap" consists of two helices--helix one spans from residues 36-44, and helix two spans from residues 48-56.
CE20
In this generation, it was found that the most catalytically efficient models had mutated T34, P48, and R56 to . These mutations further tightened the binding pocket and create a more hydrophobic environment.
Kinetics
Catalytic efficiency of 4 key generations of Diels-Alderase enzyme. Kinetic data was measured at 25 degrees Celsius, in PBS, at pH 7.4 (cite 3)
Application
The CE20 model is up to 300 fold more efficient than the first generation model, making it the most efficient Diels-Alderase yet. It surpasses many other biological (antibody) and artificial (ribozyme, metalloenzyme) attempts at catalyzing the Diels-Alder reaction. Even then, the CE20 model has a catalytic efficiency value at least 4 orders of magnitude lower than those seen in other moderately-efficient natural enzymes, demonstrating the innate slowness of the Diels-Alder reaction.
Though the rate of product formation using this enzyme is not significantly different from that found when reactants reflux free in solution, the diels-alderase shows a vast improvement in product stereoselectivity. When refluxed in a room temperature aqueous solution containing the necessary substrates, the enzyme catalyzed an over 90% conversion rate, producing only the 3R,4S endo cyclohexane product isomer. By comparison, refluxing the substrates free in solution for a similar duration of time yields a racemic (66:34) mixture of endo and exo products. It is primarily for these stereoselective benefits that this enzyme is valuable for synthetic purposes.
Future improvement of the Diels-Alderase will likely revolve around the improvement of catalytic efficiency, further constriction of the active site, and selective production of varying stereoisomers.
References
- ↑ Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St Clair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010 Jul 16;329(5989):309-13. PMID:20647463 doi:329/5989/309
Student Contributors
Taylor Donahue, Kate Thuma, Micah Zile