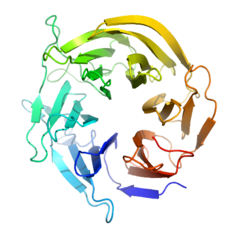We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1852
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
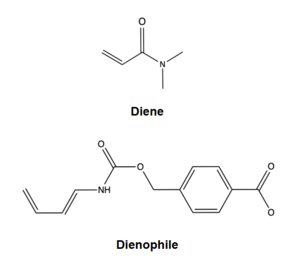
| - | [ | + | [[Image:DielsAlderasesubstrates.png|300px|left|thumb|Figure 1. Diels-Alderase Substrates]] |
| - | The Diels-Alderase protein aims to create optimal reacting conditions between the diene and dienophile in a [https://en.wikipedia.org/wiki/Diels%E2%80%93Alder_reaction Diels-Alder reaction.] It accomplishes this by decreasing the energy gap between the dienophile’s lowest unoccupied molecular orbital (LUMO) and the diene’s highest occupied molecular orbital (HOMO) in the transition state.<ref name="Siegel">PMID:20647463</ref> The binding pocket of 4O5T is selective for two substrates, 4-carboxybenzyl trans-1,3-butadiene-1-carbamate (diene) and N,N- dimethylacrylamide (dienophile). The binding site contains an H-bond donor (Y134) which lowers the LUMO energy and stabilizes the negative charge on the dienophile and an H-bond acceptor (Q208) that increases the HOMO energy and stabilizes the positive charge on the diene. Both of these H-bonding interactions work to stabilize the transition state, while also orienting the substrates in optimal conformations for reacting. | + | The Diels-Alderase protein aims to create optimal reacting conditions between the diene and dienophile in a [https://en.wikipedia.org/wiki/Diels%E2%80%93Alder_reaction Diels-Alder reaction.] It accomplishes this by decreasing the energy gap between the dienophile’s lowest unoccupied molecular orbital [https://en.wikipedia.org/wiki/HOMO_and_LUMO (LUMO)]and the diene’s highest occupied molecular orbital [https://en.wikipedia.org/wiki/HOMO_and_LUMO (HOMO)]in the transition state.<ref name="Siegel">PMID:20647463</ref> The binding pocket of 4O5T is selective for two substrates, 4-carboxybenzyl trans-1,3-butadiene-1-carbamate (diene) and N,N- dimethylacrylamide (dienophile). The binding site contains an H-bond donor (Y134) which lowers the LUMO energy and stabilizes the negative charge on the dienophile and an H-bond acceptor (Q208) that increases the HOMO energy and stabilizes the positive charge on the diene.<ref name="Siegel"/> Both of these H-bonding interactions work to stabilize the transition state, while also orienting the substrates in optimal conformations for reacting. |
The Diels-Alderase enzyme was built using de novo enzyme design, which relies on computational modeling that is refined through programming and collaborative problem-solving from online users. The original protein was made using the [https://en.wikipedia.org/wiki/Rosetta@home Rosetta] computational design program, where a potential active site was built and tested against a library of scaffold proteins. Later, as the active site was perfected, future generations of the Diels-Alderase were made using an online protein folding game called [https://en.wikipedia.org/wiki/Foldit Foldit,] where players competed to improve binding efficiency by completing various challenges. | The Diels-Alderase enzyme was built using de novo enzyme design, which relies on computational modeling that is refined through programming and collaborative problem-solving from online users. The original protein was made using the [https://en.wikipedia.org/wiki/Rosetta@home Rosetta] computational design program, where a potential active site was built and tested against a library of scaffold proteins. Later, as the active site was perfected, future generations of the Diels-Alderase were made using an online protein folding game called [https://en.wikipedia.org/wiki/Foldit Foldit,] where players competed to improve binding efficiency by completing various challenges. | ||
| + | |||
==General Structure== | ==General Structure== | ||
| - | [[Image:Squidbpropeller2.png|230px|left|thumb|Figure | + | [[Image:Squidbpropeller2.png|230px|left|thumb|Figure 1. Beta-propeller Scaffold]] |
====Scaffold==== | ====Scaffold==== | ||
After early Rosetta computational modelling, an ideal protein scaffold was found in the 6-bladed [https://en.wikipedia.org/wiki/Beta-propeller beta-propeller] of ''Loligo vulgalis,'' or the Europoean Squid. <ref name="Siegel"/><ref name="Scharff">PMID:11435114</ref> The protein is relatively simple, with only one chain, one unit, 324 residues, and no extra ligands, metal ions, or small molecules bound. | After early Rosetta computational modelling, an ideal protein scaffold was found in the 6-bladed [https://en.wikipedia.org/wiki/Beta-propeller beta-propeller] of ''Loligo vulgalis,'' or the Europoean Squid. <ref name="Siegel"/><ref name="Scharff">PMID:11435114</ref> The protein is relatively simple, with only one chain, one unit, 324 residues, and no extra ligands, metal ions, or small molecules bound. | ||
| Line 22: | Line 23: | ||
== Mechanism == | == Mechanism == | ||
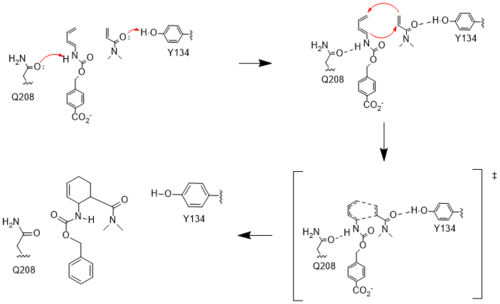
[[Image:Resizedmechanism.png|500px|left|thumb|Figure #. Active site mechanism]] | [[Image:Resizedmechanism.png|500px|left|thumb|Figure #. Active site mechanism]] | ||
| - | ==== | + | ====HOMO and LUMO==== |
The two active site residues, Y134 and Q208, use hydrogen bonding to close the energy gap between the HOMO diene and the LUMO dienophile. The goal of closing the energy gap is so that in the diels-alder reaction, the diene and dienophile can readily switch roles for the mechanism to progress and complete the formation of the product. Due to the conserved nature of this mechanism, the diels-alderase is stereoselective for the 3R, 4S endo product. | The two active site residues, Y134 and Q208, use hydrogen bonding to close the energy gap between the HOMO diene and the LUMO dienophile. The goal of closing the energy gap is so that in the diels-alder reaction, the diene and dienophile can readily switch roles for the mechanism to progress and complete the formation of the product. Due to the conserved nature of this mechanism, the diels-alderase is stereoselective for the 3R, 4S endo product. | ||
====Hydrogen Binding==== | ====Hydrogen Binding==== | ||
Rather than using acid/base catalysis like many enzymes, the diels-alderase utilized hydrogen bonding to alter the HOMO and LUMO energies of the diene and dienophile. Y134 donates a hydrogen bond to the dieneophile, increasing the electron density and lowering the LUMO. Q208 accepts a hydrogen bond from the diene, decreasing the electron density and lowering the HOMO. | Rather than using acid/base catalysis like many enzymes, the diels-alderase utilized hydrogen bonding to alter the HOMO and LUMO energies of the diene and dienophile. Y134 donates a hydrogen bond to the dieneophile, increasing the electron density and lowering the LUMO. Q208 accepts a hydrogen bond from the diene, decreasing the electron density and lowering the HOMO. | ||
| + | |||
| Line 45: | Line 47: | ||
In this generation, it was found that the most catalytically efficient models had mutated T34, P48, and R56 to <scene name='10/1075254/Ce_20_mutations/4'>I43,L48, and S56</scene>. These mutations further tightened the binding pocket and create a more hydrophobic environment. | In this generation, it was found that the most catalytically efficient models had mutated T34, P48, and R56 to <scene name='10/1075254/Ce_20_mutations/4'>I43,L48, and S56</scene>. These mutations further tightened the binding pocket and create a more hydrophobic environment. | ||
| - | + | ==Kinetics== | |
[[Image:Diels_Alderase_kinetic_table_Large.jpeg|300px|left]] | [[Image:Diels_Alderase_kinetic_table_Large.jpeg|300px|left]] | ||
Catalytic efficiency of 4 key generations of Diels-Alderase enzyme. Kinetic data was measured at 25 degrees Celsius, in PBS, at pH 7.4 (cite 3) | Catalytic efficiency of 4 key generations of Diels-Alderase enzyme. Kinetic data was measured at 25 degrees Celsius, in PBS, at pH 7.4 (cite 3) | ||
Revision as of 00:54, 16 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Diels-Alderase
| |||||||||||