We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1852
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
====HOMO and LUMO==== | ====HOMO and LUMO==== | ||
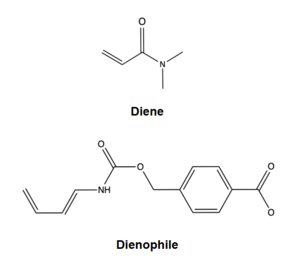
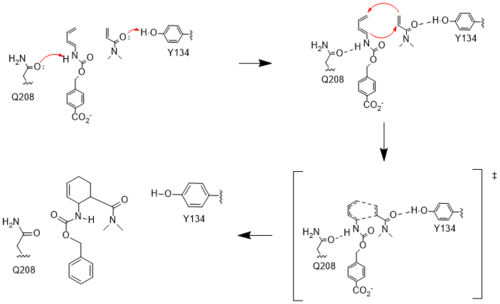
The two active site residues, Y134 and Q208, use hydrogen bonding to close the energy gap between the [https://en.wikipedia.org/wiki/Diels%E2%80%93Alder_reaction HOMO diene and the LUMO dienophile]. The goal of closing the energy gap allows the diene and dienophile to readily switch roles for the mechanism to progress and complete the formation of the product. Due to the conserved nature of this mechanism, the diels-alderase is stereoselective for the 3R, 4S endo product. | The two active site residues, Y134 and Q208, use hydrogen bonding to close the energy gap between the [https://en.wikipedia.org/wiki/Diels%E2%80%93Alder_reaction HOMO diene and the LUMO dienophile]. The goal of closing the energy gap allows the diene and dienophile to readily switch roles for the mechanism to progress and complete the formation of the product. Due to the conserved nature of this mechanism, the diels-alderase is stereoselective for the 3R, 4S endo product. | ||
| - | ====Hydrogen | + | ====Hydrogen Bonding==== |
Rather than using [https://en.wikipedia.org/wiki/Acid_catalysis acid-base catalysis]like many enzymes, the Diels-Alderase utilizes hydrogen bonding to alter the HOMO and LUMO energies of the diene and dienophile. Y134 donates a hydrogen bond to the dieneophile, increasing the electron density and lowering the LUMO. Q208 accepts a hydrogen bond from the diene, decreasing the electron density and lowering the HOMO. | Rather than using [https://en.wikipedia.org/wiki/Acid_catalysis acid-base catalysis]like many enzymes, the Diels-Alderase utilizes hydrogen bonding to alter the HOMO and LUMO energies of the diene and dienophile. Y134 donates a hydrogen bond to the dieneophile, increasing the electron density and lowering the LUMO. Q208 accepts a hydrogen bond from the diene, decreasing the electron density and lowering the HOMO. | ||
| Line 39: | Line 39: | ||
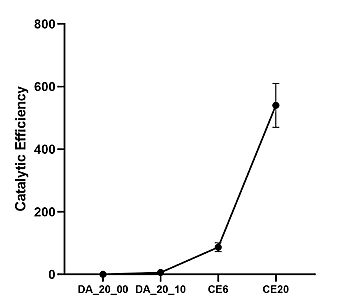
DA_20_10 provided key mutations in and around the active site that increased the hydrophobicity, provided structural stability, and increased interactions between the ligand and surrounding residues. | DA_20_10 provided key mutations in and around the active site that increased the hydrophobicity, provided structural stability, and increased interactions between the ligand and surrounding residues. | ||
=====Q162R===== | =====Q162R===== | ||
| - | + | Glu 162, a <scene name='10/1075254/Q162/2'>glutamine</scene>, resides near the top of the binding entrance to the enzyme, and is outside 3 angstroms in most models on the enzyme. It can act as a hydrogen bond donor to the terminal phosphate on the ligand when in proximity. To increase this interaction, the group chose to mutate this Q to an <scene name='10/1075254/Q_to_r/1'>arginine</scene>, which decreased the length of the potential hydrogen bond to within 2.5 angstroms, increasing the strength of the interaction.<ref name="Siegel"/> | |
=====S284A===== | =====S284A===== | ||
| - | : | + | :Ser 284 resides deep within the binding pocket of the enzyme. The group chose a <scene name='10/1075253/S284/2'>serine</scene> to <scene name='10/1075253/A285_scence/2'>alanine</scene> mutation to increase the hydrophobicity of the binding pocket and reduce reactivity, without also changing any steric characteristics in the region ''unintentionally'' near the catalytic residues.<ref name="Siegel"/> |
=====A285N===== | =====A285N===== | ||
:<scene name='10/1075254/N285/3'>N285</scene>, as follows, is also buried within the binding pocket. The group introduced this mutation to increase steric hindrance with the catalytic tyrosine, reducing the number of rotamers the residue has to increase the reactivity of the enzyme by lowering the distance between Y134 and the ligand.<ref name="Siegel"/> | :<scene name='10/1075254/N285/3'>N285</scene>, as follows, is also buried within the binding pocket. The group introduced this mutation to increase steric hindrance with the catalytic tyrosine, reducing the number of rotamers the residue has to increase the reactivity of the enzyme by lowering the distance between Y134 and the ligand.<ref name="Siegel"/> | ||
Revision as of 18:36, 17 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Diels-Alderase
| |||||||||||