We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox323
From Proteopedia
(Difference between revisions)
| Line 58: | Line 58: | ||
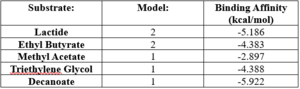
SwissDock analysis showed a preference for larger molecules, specifically fatty acids. Lactide, Ethyl Butyrate, and Triethylene Glycol exhibited noticeably weak binding affinities to the theorized active site of 4Q7Q. <ref name="SwissDockOne">Bugnon M, Röhrig UF, Goullieux M, Perez MAS, Daina A, Michielin O, Zoete V. SwissDock 2024: major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res. 2024, 52 (W1), W324-W332. DOI: 10.1093/nar/gkae300.</ref><ref name="SwissDockTwo"> | SwissDock analysis showed a preference for larger molecules, specifically fatty acids. Lactide, Ethyl Butyrate, and Triethylene Glycol exhibited noticeably weak binding affinities to the theorized active site of 4Q7Q. <ref name="SwissDockOne">Bugnon M, Röhrig UF, Goullieux M, Perez MAS, Daina A, Michielin O, Zoete V. SwissDock 2024: major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res. 2024, 52 (W1), W324-W332. DOI: 10.1093/nar/gkae300.</ref><ref name="SwissDockTwo"> | ||
| - | Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Web Server issue), W270-W277. DOI: 10.1093/nar/gkr366</ref><ref name="SwissDockThree"> | + | Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Web Server issue), W270-W277. DOI: 10.1093/nar/gkr366</ref><ref name="SwissDockThree"> Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model., 2021, 61 (8), 3891–3898, DOI: 10.1021/acs.jcim.1c00203</ref><ref name="SwissDockFour">Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem., 2010, 31 (2), 455–461, DOI: 10.1002/jcc.21334</ref> These ligands may be ill-suited to act as substrates for 4Q7Q as they are remarkably polar, and lipids—one of the potential categories of substrates for 4Q7Q—are mostly non-polar.<ref name="SwissDockOne" /><ref name="SwissDockTwo" /><ref name="SwissDockThree" /><ref name="SwissDockFour" /> |
| - | Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model., 2021, 61 (8), 3891–3898, DOI: 10.1021/acs.jcim.1c00203</ref><ref name="SwissDockFour">Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem., 2010, 31 (2), 455–461, DOI: 10.1002/jcc.21334</ref> These ligands may be ill-suited to act as substrates for 4Q7Q as they are remarkably polar, and lipids—one of the potential categories of substrates for 4Q7Q—are mostly non-polar.<ref name="SwissDockOne" /><ref name="SwissDockTwo" /><ref name="SwissDockThree" /><ref name="SwissDockFour /> | + | |
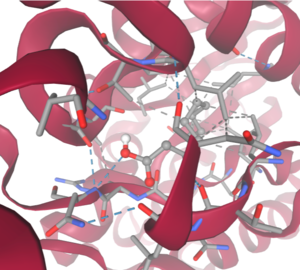
| - | [[Image:DecanoateDocking.png|300px|right|thumb|SwissDock-based analysis of the intermolecular interactions between Decanoate and 4Q7Q's proposed active site <ref name="SwissDockOne" /><ref name="SwissDockTwo" /><ref name="SwissDockThree" /><ref name="SwissDockFour />]] | + | [[Image:DecanoateDocking.png|300px|right|thumb|SwissDock-based analysis of the intermolecular interactions between Decanoate and 4Q7Q's proposed active site <ref name="SwissDockOne" /><ref name="SwissDockTwo" /><ref name="SwissDockThree" /><ref name="SwissDockFour" />]] |
Despite this, these ligands show noticeable hydrophobic interactions with the active site. This implies 4Q7Q uses hydrophobic regions to help guide substrates into the right orientation for enzymatic processes. This also further supports the possibility that 4Q7Q primarily operates with hydrophobic lipid-based substrates. This also explains why Methyl Acetate exhibited a relatively weaker affinity for 4Q7Q, as its smaller structure prevented hydrophobic interactions. | Despite this, these ligands show noticeable hydrophobic interactions with the active site. This implies 4Q7Q uses hydrophobic regions to help guide substrates into the right orientation for enzymatic processes. This also further supports the possibility that 4Q7Q primarily operates with hydrophobic lipid-based substrates. This also explains why Methyl Acetate exhibited a relatively weaker affinity for 4Q7Q, as its smaller structure prevented hydrophobic interactions. | ||
| - | [[Image:4Q7QDockingEnergies.png|300px|left|thumb|SwissDock-based analysis of the intermolecular interactions between Decanoate and 4Q7Q's proposed active site <ref name="SwissDockOne" /><ref name="SwissDockTwo" /><ref name="SwissDockThree" /><ref name="SwissDockFour />]] | + | [[Image:4Q7QDockingEnergies.png|300px|left|thumb|SwissDock-based analysis of the intermolecular interactions between Decanoate and 4Q7Q's proposed active site <ref name="SwissDockOne" /><ref name="SwissDockTwo" /><ref name="SwissDockThree" /><ref name="SwissDockFour" />]] |
=== Hypothetical Function === | === Hypothetical Function === | ||
Revision as of 21:40, 25 April 2025
4Q7Q Structure and Proposed Functionality
(NOTE TO ALL EDITORS: This page is part of a final project for a biochemistry lab at Elizabethtown College. Please do not edit this. -Neil Divins)
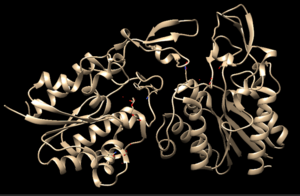
4Q7Q is a homodimeric protein complex that originates from the bacterial species Chitinophaga Pinensis and has a mass of 58.5 kDa. It is a member of the SGNH Hydrolase Superfamily with structural and sequential similarities to esterases and lipases. Current evidence suggests it causes the hydrolysis of esters and/or acetyl groups on lipids/lipid-like molecules via a serine protease-like active site.
| |||||||||||
References
- ↑ 1.0 1.1 4Q7Q. Protein Database, 2014. https://www.rcsb.org/structure/4Q7Q
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Nadzirin, N.; Gardiner, E.; Willett, P.; Artymiuk, P. J.; Firdaus-Raih, M. 2012. SPRITE and ASSAM: web servers for side chain 3D-motif searching in protein structures. Nucleic Acids Res., 40(Web Server Issue), W380-6.
- ↑ 3.0 3.1 Rio, T. G. D.; et al. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). Stand. Genomic. Sci., 2010, 2(1), 87-95. https://pmc.ncbi.nlm.nih.gov/articles/PMC3035255/
- ↑ 4.0 4.1 4.2 SGNH hydrolase superfamily. InterPro, 2017. https://www.ebi.ac.uk/interpro/entry/InterPro/IPR036514/
- ↑ 5.0 5.1 5.2 Rio, T. G. D.; et al. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). Stand. Genomic. Sci., 2010, 2(1), 87-95. https://pmc.ncbi.nlm.nih.gov/articles/PMC3035255/
- ↑ 6.0 6.1 6.2 Molgaard, A.; Kauppinen, S.; Larsen, S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Struct., 2000, 8(4), 373-383. https://www.sciencedirect.com/science/article/pii/S0969212600001180?via%3Dihub
- ↑ UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.
- ↑ Akoh, C. C.; Lee, G.; Liaw, Y.; Huang, T.; Shaw, J. GDSL family of serine esterases/lipases. Prog. Lipid Res., 2004, 43(6), 534-552. https://pubmed.ncbi.nlm.nih.gov/15522763/
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 Holm L, Laiho A, Toronen P, Salgado M (2023) DALI shines a light on remote homologs: one hundred discoveries. Protein Science 23, e4519
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedMolaard - ↑ 11.0 11.1 11.2 11.3 Bugnon M, Röhrig UF, Goullieux M, Perez MAS, Daina A, Michielin O, Zoete V. SwissDock 2024: major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res. 2024, 52 (W1), W324-W332. DOI: 10.1093/nar/gkae300.
- ↑ 12.0 12.1 12.2 12.3 Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Web Server issue), W270-W277. DOI: 10.1093/nar/gkr366
- ↑ 13.0 13.1 13.2 13.3 Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model., 2021, 61 (8), 3891–3898, DOI: 10.1021/acs.jcim.1c00203
- ↑ 14.0 14.1 14.2 14.3 Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem., 2010, 31 (2), 455–461, DOI: 10.1002/jcc.21334
- ↑ Miesfeld, R. L.; McEvoy, M. M. Biochemistry, 2nd ed.; W. W. Norton & Company, 2021
- ↑ Xia, L.; Qian, M.; Cheng, F.; Wang, Y.; Han, J.; Xu, Y.; Zhang, K.; Tian, J.; Jin, Y. The effect of lactic acid bacteria on lipid metabolism and flavor of fermented sausages. Food Biosci., 2023, 56, 103172.