We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox323
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
(NOTE TO ALL EDITORS: This page is part of a final project for a biochemistry lab at Elizabethtown College. Please do not edit this. -Neil Divins) | (NOTE TO ALL EDITORS: This page is part of a final project for a biochemistry lab at Elizabethtown College. Please do not edit this. -Neil Divins) | ||
| - | 4Q7Q is a homodimeric protein complex that originates from the bacterial species Chitinophaga Pinensis and has a mass of 58.5 kDa. It is a member of the SGNH Hydrolase Superfamily with structural and sequential similarities to esterases and lipases. Current evidence suggests it causes the hydrolysis of esters and/or acetyl groups on lipids/lipid-like molecules via a | + | 4Q7Q is a homodimeric protein complex that originates from the bacterial species Chitinophaga Pinensis and has a mass of 58.5 kDa. It is a member of the SGNH Hydrolase Superfamily with structural and sequential similarities to esterases and lipases. Current evidence suggests it causes the hydrolysis of esters and/or acetyl groups on lipids/lipid-like molecules via a catalytic triad-like active site. |
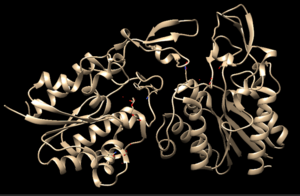
<StructureSection load='4Q7Q' size='300' side='right' caption='3D Representation of 4Q7Q's structure' scene=''> | <StructureSection load='4Q7Q' size='300' side='right' caption='3D Representation of 4Q7Q's structure' scene=''> | ||
| Line 19: | Line 19: | ||
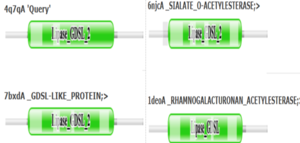
=== Family and Superfamily === | === Family and Superfamily === | ||
| - | Research shows that 4Q7Q is a member of the SGNH Hydrolase protein super family. BLAST and InterPro research suggested 4Q7Q best fits this superfamily, and the known conserved residues seen from SPRITE analysis—Serine, Glycine, Asparagine, and Histidine—line up with those observed throughout this family.<ref name="SGNH" /><ref name = "Molgaard">Molgaard, A.; Kauppinen, S.; Larsen, S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Struct., 2000, 8(4), 373-383. https://www.sciencedirect.com/science/article/pii/S0969212600001180?via%3Dihub</ref>. Notably, this superfamily is also referred to as the GDSL Hydrolase superfamily. | + | Research shows that 4Q7Q is a member of the SGNH Hydrolase protein super family. BLAST and InterPro research suggested 4Q7Q best fits this superfamily, and the known conserved residues seen from SPRITE analysis—Serine, Glycine, Asparagine, and Histidine—line up with those observed throughout this family.<ref name="SGNH" /><ref name = "Molgaard">Molgaard, A.; Kauppinen, S.; Larsen, S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Struct., 2000, 8(4), 373-383. https://www.sciencedirect.com/science/article/pii/S0969212600001180?via%3Dihub</ref>. Notably, this superfamily is also referred to as the GDSL Hydrolase superfamily.<ref name="SGNH" /><ref name="Molgaard" />. |
[[Image:4Q7QAChain.png|300px|right|thumb|Chimera-generated representation of the A chain of 4Q7Q.<ref name="Chimera">UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.</ref>]] | [[Image:4Q7QAChain.png|300px|right|thumb|Chimera-generated representation of the A chain of 4Q7Q.<ref name="Chimera">UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.</ref>]] | ||
| Line 68: | Line 68: | ||
=== Hypothetical Function === | === Hypothetical Function === | ||
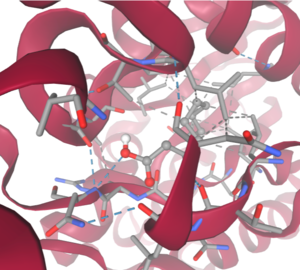
| - | Taking all of the above evidence into consideration, we currently believe 4Q7Q is an enzyme responsible for the hydrolysis of lipid esters and/or fatty acids via a | + | Taking all of the above evidence into consideration, we currently believe 4Q7Q is an enzyme responsible for the hydrolysis of lipid esters and/or fatty acids via a catalytic triad active site. This may suggest 4Q7Q plays an important role in providing energy to the body in a method similar to the beta-oxidation of fatty acids <ref name="Textbook">Miesfeld, R. L.; McEvoy, M. M. Biochemistry, 2nd ed.; W. W. Norton & Company, 2021</ref> Other studies suggest that the hydrolysis of fatty acids could be involved in fermentation-related processes or even the degradation of aryl lipid esters.<ref name="sausage">Xia, L.; Qian, M.; Cheng, F.; Wang, Y.; Han, J.; Xu, Y.; Zhang, K.; Tian, J.; Jin, Y. The effect of lactic acid bacteria on lipid metabolism and flavor of fermented sausages. Food Biosci., 2023, 56, 103172.</ref><ref name="GDSL" /> |
| - | We also believe 4Q7Q undergoes several significant structural changes during enzymatic activities. Analysis into other members of its family reveal mechanisms wherein serine and histidine residues shift during substrate binding. <ref name="GDSL" /> | + | We also believe 4Q7Q undergoes several significant structural changes during enzymatic activities. Analysis into other members of its family reveal mechanisms wherein serine and histidine residues shift during substrate binding. <ref name="GDSL" /> Specifically, the mechanism of note is similar to the ester hydrolysis or formation of lipases and esterases and is composed of four steps: First, the substrate is bound to the active serine, yielding a tetrahedral intermediate stabilized by the catalytic His and Asp residues. Next, the alcohol is released and an acyl–enzyme complex is formed. Attack of a nucleophile (water in hydrolysis) forms again a tetrahedral intermediate, which after resolution yields the product and free enzyme.<ref name="Catalytic">Bornscheuer, U. T. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev., 2002, 26(1), 73-81. https://doi.org/10.1111/j.1574-6976.2002.tb00599.x</ref> The residues involved with this catalytic mechanism are also similar to the residues located in the motif of note from SPRITE analysis.<ref name="SPRITE" /><ref name="Catalytic" /> |
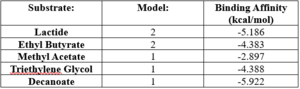
== Experimental Data == | == Experimental Data == | ||
Revision as of 20:42, 27 April 2025
4Q7Q Structure and Proposed Functionality
(NOTE TO ALL EDITORS: This page is part of a final project for a biochemistry lab at Elizabethtown College. Please do not edit this. -Neil Divins)
4Q7Q is a homodimeric protein complex that originates from the bacterial species Chitinophaga Pinensis and has a mass of 58.5 kDa. It is a member of the SGNH Hydrolase Superfamily with structural and sequential similarities to esterases and lipases. Current evidence suggests it causes the hydrolysis of esters and/or acetyl groups on lipids/lipid-like molecules via a catalytic triad-like active site.
| |||||||||||
References
- ↑ 1.0 1.1 4Q7Q. Protein Database, 2014. https://www.rcsb.org/structure/4Q7Q
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Nadzirin, N.; Gardiner, E.; Willett, P.; Artymiuk, P. J.; Firdaus-Raih, M. 2012. SPRITE and ASSAM: web servers for side chain 3D-motif searching in protein structures. Nucleic Acids Res., 40(Web Server Issue), W380-6.
- ↑ 3.0 3.1 Rio, T. G. D.; et al. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). Stand. Genomic. Sci., 2010, 2(1), 87-95. https://pmc.ncbi.nlm.nih.gov/articles/PMC3035255/
- ↑ 4.0 4.1 4.2 SGNH hydrolase superfamily. InterPro, 2017. https://www.ebi.ac.uk/interpro/entry/InterPro/IPR036514/
- ↑ 5.0 5.1 5.2 Rio, T. G. D.; et al. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). Stand. Genomic. Sci., 2010, 2(1), 87-95. https://pmc.ncbi.nlm.nih.gov/articles/PMC3035255/
- ↑ 6.0 6.1 6.2 6.3 Molgaard, A.; Kauppinen, S.; Larsen, S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Struct., 2000, 8(4), 373-383. https://www.sciencedirect.com/science/article/pii/S0969212600001180?via%3Dihub
- ↑ UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.
- ↑ Akoh, C. C.; Lee, G.; Liaw, Y.; Huang, T.; Shaw, J. GDSL family of serine esterases/lipases. Prog. Lipid Res., 2004, 43(6), 534-552. https://pubmed.ncbi.nlm.nih.gov/15522763/
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 Holm L, Laiho A, Toronen P, Salgado M (2023) DALI shines a light on remote homologs: one hundred discoveries. Protein Science 23, e4519
- ↑ 10.0 10.1 10.2 10.3 Bugnon M, Röhrig UF, Goullieux M, Perez MAS, Daina A, Michielin O, Zoete V. SwissDock 2024: major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res. 2024, 52 (W1), W324-W332. DOI: 10.1093/nar/gkae300.
- ↑ 11.0 11.1 11.2 11.3 Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Web Server issue), W270-W277. DOI: 10.1093/nar/gkr366
- ↑ 12.0 12.1 12.2 12.3 Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model., 2021, 61 (8), 3891–3898, DOI: 10.1021/acs.jcim.1c00203
- ↑ 13.0 13.1 13.2 13.3 Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem., 2010, 31 (2), 455–461, DOI: 10.1002/jcc.21334
- ↑ Miesfeld, R. L.; McEvoy, M. M. Biochemistry, 2nd ed.; W. W. Norton & Company, 2021
- ↑ Xia, L.; Qian, M.; Cheng, F.; Wang, Y.; Han, J.; Xu, Y.; Zhang, K.; Tian, J.; Jin, Y. The effect of lactic acid bacteria on lipid metabolism and flavor of fermented sausages. Food Biosci., 2023, 56, 103172.
- ↑ 16.0 16.1 Bornscheuer, U. T. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev., 2002, 26(1), 73-81. https://doi.org/10.1111/j.1574-6976.2002.tb00599.x