Overview
Through the 2024-2025 Spring Semester at Elizabethtown College, analysis of protein 2O14 was performed to determine its enzymatic function and protein class. Determining the function of unknown proteins is beneficial to help understand biological processes. For 2O14 in particular, this protein was found in Bacillus subtilis from soil and the gastoinestrial tract of ruminants and humans. Understanding its enzymatic function will pinpoint exactly how this organism interacts with the environment and humans.
2O14 exists as monomeric protein complex. Using SPRITE and Chimera revealed one chain that is very long and does not replicate the amino acid chain. This was further confirmed by looking at the X-ray structure of protein 2O14 and it is stating that there is only one unique protein chain. The chain is 366 residues long and the entire complex has a molecular weight of approximately 41.79kDa.
2O14 proteins originated from the bacterial species bacillus subtills. InterPro results showed how almost all similar enzymes came from bacterial species like the bacillus subtills. It was also found to not be in any viruses, but a small fraction could be found in archaea, eukaryote and other small organisms.
Family and Superfamily
The data found showed that 2O14 is like a Rhamnogalacturan_acetylesterase that falls under the SGNH Hydrolase protein super family. Using a SPRITE analysis, we found that the lower RMSD values match was with 1bwp_c01 a Platelet Act Factor with a value of 0.60 and matched with aspargine, aspartic acid and histidine residues, for the right-hand superposition. Looking at the left-hand superposition, the lowest RMSD value was 0.62 with 1xny_c01, propionyl-COA carboxylase complex B, and its matching residues were 1 alanine and two glycine groups.
Using the data gotten from the DALI software, it was found that protein 1k7c or rhamnogalacturonan acetylesterase had the most matching amino acids, 98, creating a z score pf 25.1 create a 25.1% match to protein 2O14. Furthermore, using DALI stacked Pfam data graphics showed that protein 2O14 was a potential member of GDSL-like Lipase/Acylhyydrolase family. This would agree with data gather from InterPro that stated that 2O14 was most likely resembling a rhamnogalacturonan acetylesterase.
When using BLAST, the group was able to not only look at 2O14 amino acid sequence but also its taxonomy which showed that the protein was seen in the Bacillota superfamily which is a phylum of bacteria that have gram positive cell walls. Furthermore, for this phylum of bacteria it is very common for them to contain multiple hydrolases including SGNN hydrolases like 2O14.
Sequence Analysis
Analysis of the primary structure of 2O14 began in SPRITE. Right-handed superposition analysis related two possible enzymes to 2O14, 1BWP and 1PP4. RMSD values for 1BWP and 1PP4 were 0.92 and 0.93 respectively and both proteins matched five residues with 2O14. 1BWP is defined as “Platelet-activating factor acetyl hydrolase,” and 1PP4 is defined as “Rhamnogalacturonan acetyl esterase.”
The primary structure of 2O14 exhibits its active site through the specific residues of Ser171, Gly209, Asn241, Asp339, and His342. This active site spans residues 170 to 350 with this region defined as “Rhamnogalacturonan acetyl esterase like” and falling into the SGNH hydrolase superfamily through BLAST analysis of the primary structure.
Analysis of the primary sequence alone showed promising signs of 2O14 acting as a hydrolase due to its relation to RGAE through its active site and the SGNH hydrolase superfamily.
Structural Analysis
2O14 contains several similarities secondary structures to proteins 1K7C and 1PP4. 2O14 exhibits a GDSL 2 sequence motif, a variant of the GDSL motif exhibited in both 1K7C and 1PP4. Structural analysis using the Dali software showed both proteins superimposed over 2O14 with z-scores of 25.1 and 24.8 respectively. These two proteins are defined as rhamnogalacturonan acetyl esterases.
After obtaining data from SPRITE, 2O14 was superimposed onto 1PP4 giving an angstroms difference of 0.736Å. This mostly came in the alpha helix region both proteins where the proposed active site is located. Through all analyses, the beta sheet region of 2O14 had no hits relating to its structure or sequence.
Analysis of the structure further emphasized the similarities to RGAE and the proposed function of 2O14 being a hydrolase of some kind, either a lipase or esterase.
Substrates
SwissDock analysis was done on specific substrates including 4-Nitrophenyl alpha-glucoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, and p-nitrophenyl phosphate. Both PNPG and GlcNac exhibited high binding affinities to 2O14. This would be due 2O14’s proposed enzymatic function.
Theoretical Functionality and Proposed Bodily Purpose
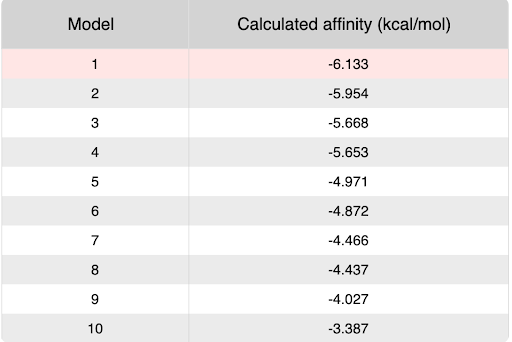
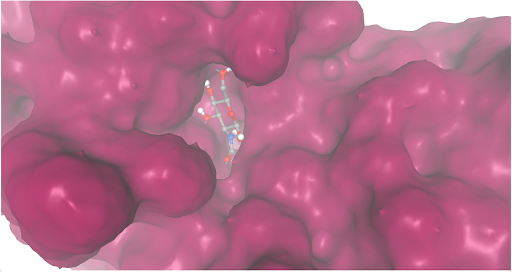
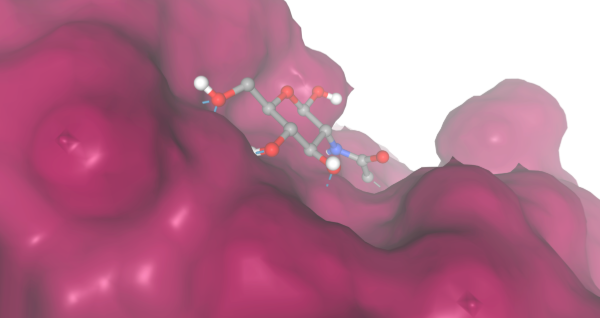
During our research and experiments the group had used SwissDock to potentially bind to protein 2O14. The determined smile code for the protein was C1=CC(=CC=C1[N+](=O)[O-])O[C@@H]2[C@@H]([C@H]([C@@H]([C@H](O2)CO)O)O)O, and found relatively low calculated affinity values ranging from -6.133 to -3.387. The SwissDock predicted four binding sites it believed molecules could interact with the protein at. One inside the protein itself as shown in Figure 1. Other spaces that were seen to have good binding affinity were outside of the protein embedded in the side, Figure 2, And then finally on the side in Figure 3. The two more likely cases of binding are figures 2 and 3, however, figure 1 had the lowest binding affinity.
Each of the binding affinities correlated with some kind of saccharide, each saccharide was found to interact with acetyl esterases. Furthermore, all the prior data had shown that protein 2O14 was most like an acetyl esterase being most closely related to Rhamnogalacturan acetylesterase, the group decided to experiment on the protein using PNPP to see whether their protein interacted with it.
After performing all the necessary steps to extract the protein, it was found that most of the protein came out in the first elution, elution 1. Then using UV-Vis, 10µL of protein solution was added to a cuvette with a 10:1 ratio of PNPP and PNPP buffer solution. This showed an instantly yellow solution, so it was believed that protein was in the elution. We did the same test again with elution 2, to confirm that the protein was in elution 1, and had similar results. The group then attempted to adjust the pH to be more like dirt and soil pH and got completely opposite results, where the absorbance of the first two tests was around 0.95 and the pH test was around 0.09. With these completely opposite results, it was unclear if elution 1 had the protein. Figures 4, 5, and 6 show the explained experiments above in order.
Protein Extraction
The protein was grown inside of E.coli with a His-tag for resistance to the antibiotic, Ampicillin. Once the protein was grown, the protein solution was centrifuged and sonicated multiple times to extract the expressed protein. For purification, the protein solution was ran through a Ni-NTA column gathering approximately 5 mL of protein elutions in total.
Figure 4
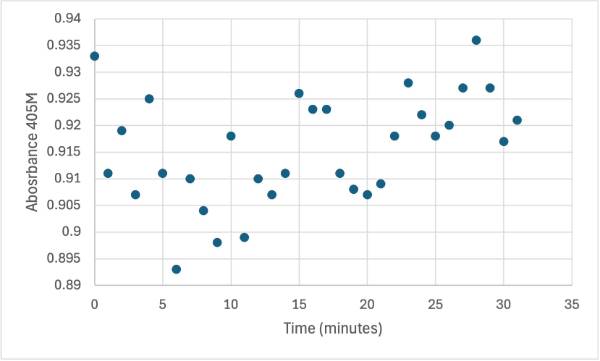
Protein test using 10µL of protein from elution and 30mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M.
Figure 5
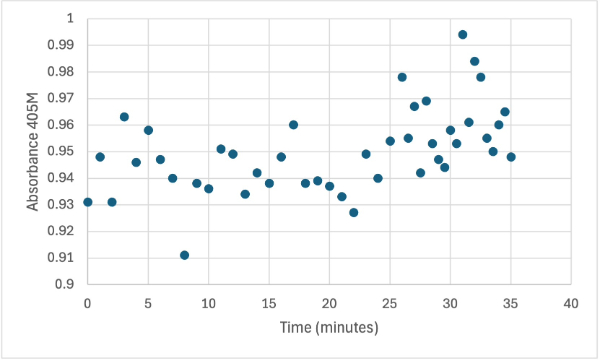
Protein test using 10µL of protein from elution 2 and 30mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M.
Figure 6
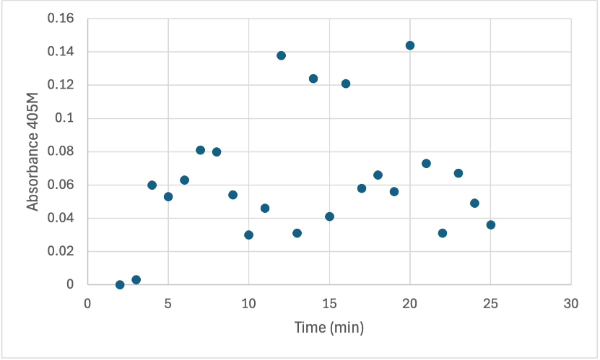
Protein test using 10µL of protein from elution 1 and 30mg of PNPP and 3mL of PNPP solution, changing the pH to about 5.60. Measuring absorbance at 405M.
Figure 7
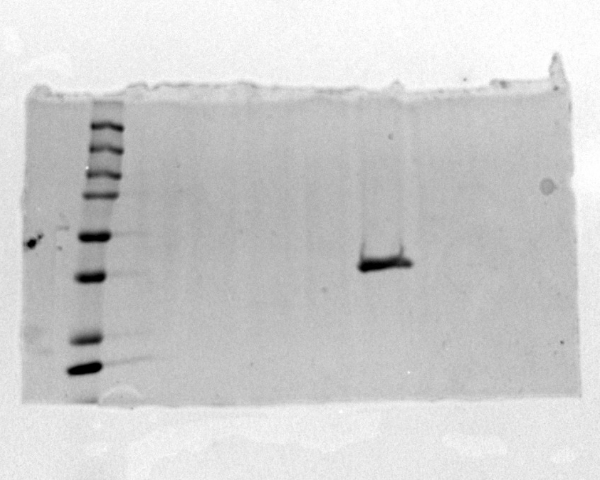
Image of SDS-PAGE gel that proves that protein was present in the sample.
The following week two more experiments were run, one with a 1:1 ratio of PNPP and PNPP solution with 10µL of protein solution for elution 1. The first was just with the PNPP mixture and showed no positive results, with absorbance values bouncing back and forth between values in the range of 0.15 to 0.096. After this test, a drop of 3.0M HCL was added to the solution to increase acidity slightly, which harbored almost identical results to the previous test. After, an experiment with PNPA was performed to see if a change in substrate would cause the protein to react. Figures 8, 9, and 10 show the results of these reactions.
Figure 8
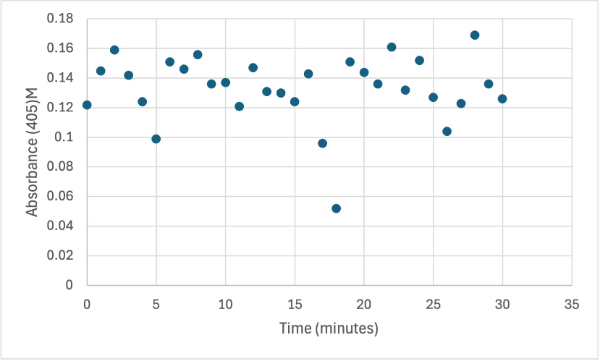
Protein test using 10µL of protein from elution 1 and 3mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M.
Figure 9
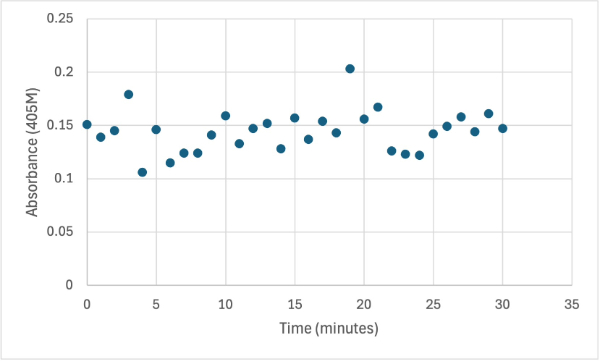
Protein test using 10µL of protein from elution 1 and 3mg of PNPP and 3mL of PNPP solution and 1 drop of HCL with a pH of about 10.50. Measuring absorbance at 405M.
Figure 10
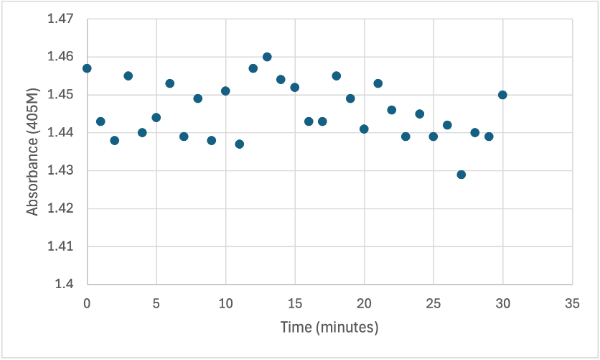
Protein test using 10µL of protein from elution 1 and 10mg of PNPA and 10mL of PNPP solution and 1 drop of HCL with a pH of about 10.50. Measuring absorbance at 405M.
The following week two more reactions were run to prove that the protein function was as an acetyl esterase. The second to last experiment was run with a ratio for 5:1 PNPP and PNPP solution which resulted in no absorbance change throughout the experiment. Then as a final used PNPA with a 5:1 ratio solution, and during this experiment 5µL of protein solution was added to the cuvette solution being measured. This was done to see if more protein concentration was needed to perform the reaction, and after 30 minutes and 155µL of protein solution, no absorbance change was observed. Figures 11 and 12 show these reactions.
Figure 11
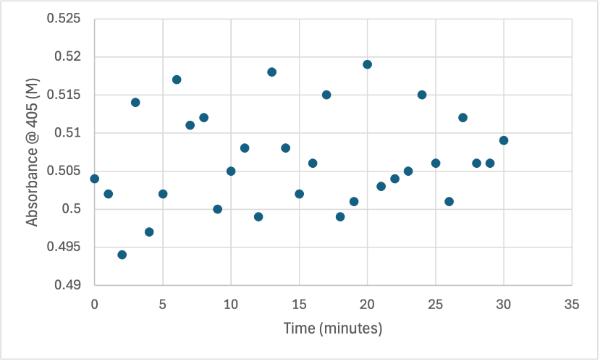
Protein test using 10µL of protein from elution 1 and 15mg of PNPA and 3mL of PNPP solution. Measuring absorbance at 405M.
Figure 12
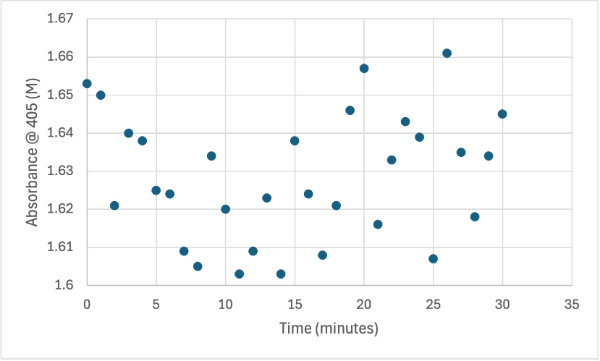
Protein test with increasing protein solution of 5µL every minute from elution 1 with 15mg of PNPA and 3mL PNPP solution. The pH of the solution is 8.35 and it was measuring absorbance at 405M.
This data shows two possible case results. One is that the protein sample might have been out of a cold environment for too long which would cause the protein to denature and not operate as predicted. Another potential case is that the predicted function of the protein was incorrect.