Introduction
SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) is an RNA virus that is responsible for the highly infectious respiratory disease, COVID-19. It first emerged late in 2019 and quickly spread around the world, earning the rank of "pandemic" by March of the following year. Given the severity of the situation the entire world was in, researchers around the globe began researching the virus and methods to prevent its spread. One such method looked to stop the initial infection by preventing the endocytosis of the virus itself through direct competition (for the virus) with the receptor to which it binds. Given this is a respiratory infection, the mode by which to give this competitive inhibitor was designed to be a nose-spray. Initial research looked into the use of antibodies to accomplish this task but found antibodies were not stable enough for this mode. But the use of a new tool changed the game. Using a protein-predicting software, generated proteins, known as miniproteins or mini binders due to their relatively small size, solved the stability issue and greatly increased binding affinity. Not only has this tool led to a major step forward in the treatment of COVID-19, but it can even be applied to other infections, marking a new era in the development of therapeutics.
Spike Protein
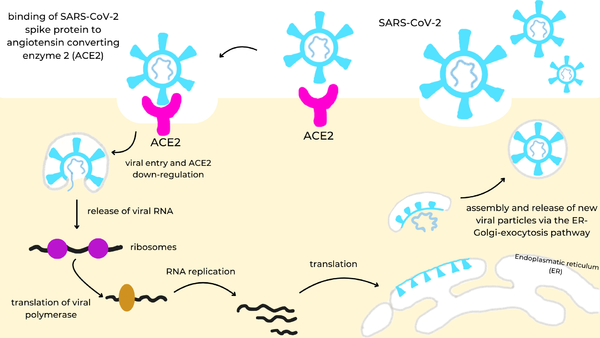
To begin, the spike protein[1] is a homotrimer glycoprotein that lies on the surface of SARS-CoV-2 to facilitate the entry of the virus into host cells. The spike utilizes the Angiotensin Converting Enzyme Receptor 2 (ACE2)[2] in the lungs to enter. Once inside the cell, virus-specific RNA and proteins are synthesized within the cytoplasm. The viral envelope merges with the oily membrane of our own cells, allowing the virus to release its genetic material into the inside of the healthy cell. Each infected cell may produce and release millions of copies of the virus which can infect other neighboring cells and people when the viral particles are released from the airways (i.e., via coughing or sneezing). This pathway is visualized in the image to the right.
Receptor Binding Domains
The spike protein contains three Receptor Binding Domains , containing the binding site (one per chain). A at the RBDs reveal a mostly beta-sheet secondary structure with a handful of helices scattered about. There are also a couple variable loops where a number of interactions take place as well. These interactions are specific to the binding ligand and will be talked about in depth later.
ACE2 Receptor
the natural receptor for the spike protein, is a membrane-bound protein located in many parts of the body. It is made up of 805 amino acids and its primary role is to be a part of the regulation of blood pressure. Its binding site is located within a single extracellular N-terminal domain. Residues 23-46 in this region form an alpha helix which makes the majority of with the RBD of the spike protein. These interactions are stabilized by hydrogen bonds[3] and Van der Waals forces[4].
Mini Protein Inhibitors
AHB2
is one of the miniproteins that was tested. It was manually developed by taking the binding helices in ACE2 (residues 23-46) and making individual changes to better binding affinity. This molecule utilizes Van der Waals interactions in its hydrophobic core to stabilize. show an increased number of interactions between the spike and minibinder. This allows it to compete with the ACE2 receptor more effectively, preventing it from binding with the spike protein.[1]
LCBs
The "LCBs" are the fully de novo minibinders that were generated by the protein-predicting software using information from both ACE2 and the spike protein as guidelines, one of which being . In comparison to AHB2, it is more compact and interactions with the RBD have been optimized. Its show a large number of hydrogen bonds as well as a small hydrophobic effect to promote stronger binding.
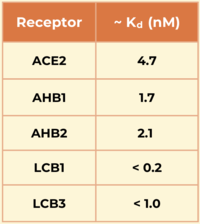
Another miniprotein, , was developed using the same computational method as LCB1 and binds to the same region of the RBD, except in the opposite direction. Its also show a large number of hydrogen bonds. However, LCB1 has more surface area and fits more precisely with the spike protein, giving it a slightly higher binding affinity (shown in the table to the right).
Significance
The researchers demonstrated the advantages of using a de novo approach to design efficient miniprotein inhibitors that bind the SARS-CoV-2 spike protein with exceptionally high affinity. The small, hyperstable miniproteins that were created using this approach (i.e., LCB1 and LCB3) are smaller, more stable, cheaper to produce, and better suited for delivery (e.g., nasal sprays) compared to traditional antibodies. By targeting the receptor-binding domain (RBD) and competing directly with ACE2, these inhibitors block viral entry into host cells, as they bind over 50 times more tightly than the ACE2 receptor.
The final structures of LCB1 and LCB3 differed by around had very low deviation (~ 1.3-1.9 Å) from their computer models, demonstrating the accuracy of the computational tools used in predicting the three-dimensional folding of the miniproteins along with their specific interactions with the spike RBD.
Therapeutic Relevance
The LCBs have a significantly higher affinity for the spike RBD and are smaller and more stable than traditional antibodies. Their reduced size allows these molecules to tightly pack more interactions in the active site as well as improves their ability to enter the respiratory system. Their hyperstability makes them able to withstand high and low temperatures, allowing them to be stored anywhere--contributing to their higher efficacy than most traditional antibodies. Overall, the development of LCB1 and LCB3 validates the reliability, efficiency, and accuracy of the computational design approach, demonstrating a quick and efficient strategy for producing therapeutics in response to emerging infectious diseases.
[2]
[3]
[4]
[2]
[3]
References
- ↑ Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park YJ, Strauch EM, Stewart L, Diamond MS, Veesler D, Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020 Oct 23;370(6515):426-431. PMID:32907861 doi:10.1126/science.abd9909
- ↑ 2.0 2.1 Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ 3.0 3.1 Gusev E, Sarapultsev A, Solomatina L, Chereshnev V. SARS-CoV-2-Specific Immune Response and the Pathogenesis of COVID-19. Int J Mol Sci. 2022 Feb 2;23(3):1716. PMID:35163638 doi:10.3390/ijms23031716
- ↑ Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 Mar 30. pii: 10.1038/s41586-020-2180-5. doi:, 10.1038/s41586-020-2180-5. PMID:32225176 doi:http://dx.doi.org/10.1038/s41586-020-2180-5
Student Contributors
- Carson Powers
- Rachael Vavul
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.