Journal:Acta Cryst F:S2053230X25007034
From Proteopedia
(Difference between revisions)

| Line 10: | Line 10: | ||
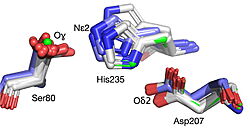
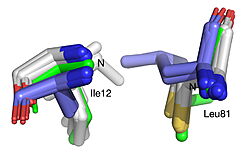
The 1.99-Å crystal structure of HNL6V reveals the molecular basis for this engineered transformation. The seven substitutions induced <scene name='10/1087242/Ca_dist_HNL6V_to_HbHNL_all/1'>subtle but systematic shifts throughout the protein</scene>, moving the catalytic atoms 0.2-0.8 Å closer to their positions in the target esterase while increasing flexibility in the lid domain—demonstrating that the mutations both repositioned key atoms and fine-tuned protein dynamics. | The 1.99-Å crystal structure of HNL6V reveals the molecular basis for this engineered transformation. The seven substitutions induced <scene name='10/1087242/Ca_dist_HNL6V_to_HbHNL_all/1'>subtle but systematic shifts throughout the protein</scene>, moving the catalytic atoms 0.2-0.8 Å closer to their positions in the target esterase while increasing flexibility in the lid domain—demonstrating that the mutations both repositioned key atoms and fine-tuned protein dynamics. | ||
| - | [[Image: | + | [[Image:019 Fig 3a.triad label.jpg|thumb|left|250px|Displacement of Cα atoms (ΔCα) in HbHNL (PDB entry [[1yb6]]), HNL40 (PDB entry [[8sni]]), and HNL71 (PDB entry [[9clr]]) relative to wt SABP2 (PDB entry [[1y7i]]). ]] [[Image:019_Fig_3b.oxyanion_label.jpg|thumb|right|250px|Displacement of Cα atoms (ΔCα) in HbHNL (PDB entry [[1yb6]]), HNL40 (PDB entry [[8sni]]), and HNL71 (PDB entry [[9clr]]) relative to wt SABP2 (PDB entry [[1y7i]]). ]] |
Revision as of 17:56, 11 August 2025
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.